¶ Introduction
The chapter gives detailed insights into the components and processes of Sumo models. This information helps the user to understand the interactions between components, indicates the model logic and consideration behind the mathematical description.
¶ Components in Sumo models
There are five symbols that distinguish the types of components, also referred to as state variables (SVs):
- soluble (S),
- colloidal (C),
- particulate (X),
- gaseous (G),
- and enthalpy (H).
The soluble SVs are transported through water, and colloidal SVs and particulate SV become part of the sludge which then can be separated from water through the settling process.
| Components | ||||
|---|---|---|---|---|
| Symbol | Name | Model | Definition | Unit |
| SVFA | Volatile fatty acids (VFA) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A fermented product representing a combination of acetate and other volatile fatty acids produced in an anaerobic environment from SB. It is available for biological removal by OHOs, CASTOs (PAOs, GAOs), and AMETOs and chemical removal during HFO reduction. | g COD.m-3 |
| SB | Readily biodegradable substrate (non-VFA) | Mini_Sumo, Sumo1, Sumo2, Sumo2S, Sumo4N | Non-VFA organic material that can be fermented to VFA, it represents a group of readily biodegradable organic material present in wastewater. | g COD.m-3 |
| SB,mono | Readily biodegradable substrate as monomers (non-VFA) | Sumo2C | Small molecular weight substrate mainly consumed by AHOs in 1st stage of high-rate process. | g COD.m-3 |
| SB,poly | Readily biodegradable substrate as polymers | Sumo2C | Readily biodegradable substrate that is not degraded in 1st stage of a high-rate process. Consumed by OHOs in 2nd stage. | g COD.m-3 |
| SMEOL | Methanol (MEOL) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A methyl alcohol or the simplest alcohol. A commonly used external carbon source for denitrification. | g COD.m-3 |
| CB | Colloidal biodegradable substrate | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A biodegradable organic component, flocculates to form XB. Analytically can be approximated by flocculation or filtration (larger than 0.1 micron but smaller than 1.2 micron) | g COD.m-3 |
| XB | Slowly biodegradable substrate | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N |
High molecular weight particulates that are hydrolyzed by extracellular enzymes to release SB. They are introduced directly from the influent. In Mini_Sumo they are also released during the bacterial decay process in death-regeneration concept models. |
g COD.m-3 |
| XB,e | Slowly biodegradable substrate from biomass decay | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N |
High molecular weight particulates that are hydrolyzed by extracellular enzymes to release SB, with a lower rate than XB. Tehy are released during the bacterial decay process in death-regeneration concept models.
|
g COD.m-3 |
| SU | Soluble unbiodegradable organics | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A soluble organic material that is not degraded biologically or chemically in wastewater and leaves in the effluent. Typically, measured by performing a flocculated filtration test on the effluent of a nitrifying plant. It also has a nitrogen and phosphorus content. There is only one process, the Thermal Hydrolysis Process, which generates SU from XU in the model. | g COD.m-3 |
| CU | Colloidal unbiodegradable organics | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A non-biodegradable organic material, flocculates to form XU and leaves the plant in the WAS or cake. Analytically can be approximated by flocculation or filtration (larger than 0.1 micron but smaller than 1.2 micron) | g COD.m-3 |
| XU | Particulate unbiodegradable organics | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A non-biodegradable organic material that is larger than 1.2 micron. It is not hydrolysed and remains untransformed due to biological and chemical reactions. The thermal hydrolysis process model is the only unit that converts a portion of XU to SU. | g COD.m-3 |
| XSTO | Storage product of AHOs | Sumo2C | An internal cell storage organic material stored by AHOs in low SRT systems using SB,mono and SVFA. Its production doesn’t involve any energy and growth. | g COD.m-3 |
| XPHA,PAO | Polyhydroxyalkanoates (PHA) stored by PAOs | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Poly-hydroxy alkanoates considered as an internal organic cellular storage product of CASTOs (PAOs). The composition of alkanoates is represented as poly-β-hydroxybutyrate. | g COD.m-3 |
| XPHA,GAO | Polyhydroxyalkanoates (PHA) stored by GAOs | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Poly-hydroxy alkanoates considered as an internal organic cellular storage product of CASTOs (GAOs). The composition of alkanoates is represented as poly-β-hydroxybutyrate. | g COD.m-3 |
| XE | Endogenous decay products | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | An organic material released during cell lysis under aerobic and anoxic environment, it has an extremely slow conversion rate of 0.07 per day and it converts to XB while releasing SNHx and SPO4. The XE builds in a system with increasing SRT. | g COD.m-3 |
| XE,ana | Anaerobic endogenous decay products | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | An organic material released on cell lysis under anaerobic environment and hydrolysed in aerobic environment to release SB, SNHx, and SPO4. This component is responsible for the additional VS destruction observed in a post aerobic digestion process. | g COD.m-3 |
| XOHO | Ordinary heterotrophic organisms (OHO) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Generalist facultative ordinary hetrotrophic organisms that consume different soluble biodegradable organics including SB, SVFA, and XMEOL and can perform biological removal under aerobic, anoxic, and anaerobic environments. They are also responsible for hydrolysis of the particulates. In MiniSumo and Sumo1, they perform one step denitrification, meaning from SNOx to SN2. In other models they follow two steps, SNO3 reduction to SNO2, and SNO2 reduction to SN2. | g COD.m-3 |
| XCASTO | Carbon storing organisms (CASTO) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A special group of carbon storing heterotrophic organisms representing a combination of both PAOs and GAOs. The process conditions dictate the ratio of PAO to GAO achieved in the process model. They are responsible for taking part in the EBPR process. They store PHA and/or GLY during the anaerobic conditions, and consume stored carbon during anoxic and aerobic environments to generate polyphosphate (XPP). In Sumo1, they perform one step denitrification, meaning from SNOx to SN2. Is other models they follow two steps, SNO3 reduction to SNO2, and SNO2 reduction to SN2. | g COD.m-3 |
| XMEOLO | Anoxic methanol utilizers (MEOLO) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Specialist heterotrophic organisms responsible for removal of XMEOL. Only found in systems with methanol addition and compete with OHOs during denitrification process. In Sumo1, they perform one step denitrification, meaning from SNOx to SN2. Is other models they follow two steps, SNO3 reduction to SNO2, and SNO2 reduction to SN2. | g COD.m-3 |
| XAHO | Carbon adsorption heterotroph organisms (AHO) | Sumo2C | Heterotrophic organisms storing readily biodegradable small molecular weight components represented as SB,mono and volatile fatty acids (SVFA) into XSTO. The growth rate of AHOs is higher than that of the OHOs and outgrow them in a short SRT system of <2 days. They only carry out aerobic consumption of the XSTO and are expected to be seeded from the influent. | g COD.m-3 |
| XNITO | Aerobic nitrifying organisms (NITO) | Mini_Sumo, Sumo1 | Obligate aerobic autotrophic organism responsible for complete nitrification, from SNHx to SNOx. It represents a combination of AOBs and NOBs for simplification in the referred models. | g COD.m-3 |
| XAOB | Aerobic ammonia oxidizers (AOB) | Sumo2, Sumo2C, Sumo2S, Sumo4N | Obligate aerobic autotrophic organism responsible for the first step in nitrification, from SNHx to SNO2. | g COD.m-3 |
| XNOB | Nitrite oxidizers (NOB) | Sumo2, Sumo2C, Sumo2S, Sumo4N | Obligate aerobic autotrophic organism responsible for the second step in nitrification, from SNO2 to SNO3. | g COD.m-3 |
| XAMX | Anammox organisms (AMX) | Sumo2, Sumo2C, Sumo2S, Sumo4N | A chemoautotrophic anaerobic bacteria that oxidizes ammonium with nitrite as the electron acceptor and with CO2 as the main carbon source. They are slow growers and have a long doubling time. | g COD.m-3 |
| XAMETO | Acidoclastic methanogens (AMETO) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Anaerobic archea that consume SVFA to produce SCH4 and SCO2 as a metabolic by-product and don’t grow under aerobic conditions. | g COD.m-3 |
| XHMETO | Hydrogenotrophic methanogens (HMETO) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Anaerobic archea that consume SH2 and SCO2 to produce SCH4 as a metabolic by-product and don’t grow under aerobic conditions. | g COD.m-3 |
| XASRO | Acidoclastic sulfate-reducing organisms (ASRO) | Sumo2S | A group of bacteria and archaea that perform anaerobic respiration utilizing SSO4 and SVFA, reducing it to SH2S and generating SCO2. They compete with AMETOs for SVFA and negatively impact performance of a digester. | g COD.m-3 |
| XHSRO | Hydrogenotrophic sulfate-reducing organisms (HSRO) | Sumo2S | A group of bacteria and archaea that perform anaerobic respiration utilizing SSO4 and SH2, reducing it to SH2S. They compete with HMETOs for SH2 consumption and negatively impact performance of a digester. | g COD.m-3 |
| XSOO | Sulfur-oxidizing organisms (SOO) | Sumo2S | They oxidize SH2S in two steps under aerobic and anoxic environments, first from SH2S to XS (elemental sulfur) and second from XS to SSO4. | g COD.m-3 |
| XALGAE | Photosynthetic organisms (ALGAE) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A group of photosynthetic organisms which grow, assimilate nutrients, SCO2 and generate SO2 in the presence of light. They consume SO2 through respiration. | g COD.m-3 |
| SNHx | Total ammonia (NHx) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Total ammonia nitrogen is a sum of ammonium and free ammonia. An essential nutrient for the biokinetic growth reactions. | g N.m-3 |
| SNH2OH | Hydroxylamine (NH2OH) | Sumo4N | An intermediate product produced by AOB from SNHx. It is then oxidized to SNO under aerobic environments. It is also reduced to form SN2O under anoxic environments. | g N.m-3 |
| SNOx | Nitrate and nitrite (NOx) | Mini_Sumo, Sumo1 | A sum of nitrate and nitrite, and electron acceptor for anoxic reactions. In the referred Models it is considered nitrate for all COD conservation purposes. | g N.m-3 |
| SNO2 | Nitrite (NO2) | Sumo2, Sumo2C, Sumo2S, Sumo4N | A sum of nitrous acid and nitrate ion and electron acceptor for anoxic reactions. | g N.m-3 |
| SNO3 | Nitrate (NO3) | Sumo2, Sumo2C, Sumo2S, Sumo4N | An electron acceptor for anoxic reactions. | g N.m-3 |
| SNO,AOB | Nitric oxide of AOB (NO) | Sumo4N | Nitric oxide generated by AOBs from SNH2OH and either oxidized to SNO2 and reduced to SN2O. It is considered as an internal product. | g N.m-3 |
| SNO,OHO | Nitric oxide of OHO (NO) | Sumo4N | Nitric oxide generated by OHOs during SNO2 reduction. It is considered as an internal product. | g N.m-3 |
| SN2O | Nitrous oxide (N2O) | Sumo4N | Generated by OHOs from SNO,OHO and by AOBs from SNO,AOB. | g N.m-3 |
| SN2 | Dissolved nitrogen (N2) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Only nitrogenous product of denitrification except for in Sumo4N model and is subject to gas transfer. | g N.m-3 |
| SN,B | Soluble biodegradable organic N (from SB) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A nitrogen containing soluble organic material (urea, amino acids, amines, and others) that releases nitrogen as total ammonia on ammonification in wastewater treatment. In the model it doesn’t have COD associated. | g N.m-3 |
| XN,B | Particulate biodegradable organic N (from XB) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A nitrogen containing particulate organic material (proteins and others) that releases SN,B on hydrolysis in wastewater treatment. In the model it doesn’t have COD. | g N.m-3 |
| XN,U | Particulate unbiodegradable organic N | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A nitrogen containing particulate organic material that is not biologically or chemically degraded in the plant and leaves with the cake solids. The only process that degrades XN,U to SNHx is the thermal hydrolysis process. | g N.m-3 |
| SPO4 | Orthophosphate (PO4) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Soluble inorganic phosphorus, represents a combination of Phosphoric acid, Dihydrogen phosphate ion, Hydrogen phosphate ion, Phosphate ion. An essential nutrient for the biokinetic growth reactions. | g P.m-3 |
| XPP | Stored polyphosphate (PP) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Poly-phosphate considered as an internal inorganic cellular storage product of CASTOs (PAOs). The composition of XPP is (Ca0.1K0.1Mg0.35PO3)n. | g P.m-3 |
| SP,B | Soluble biodegradable organic P (from SB) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A phosphorus containing soluble organic material that releases phosphorus as SPO4 in wastewater treatment. In the model it doesn’t have COD. | g P.m-3 |
| XP,B | Particulate biodegradable organic P (from XB) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A phosphorus containing particulate organic material that releases SP,B on hydrolysis in wastewater treatment. In the model it doesn’t have COD. | g P.m-3 |
| XP,U | Particulate unbiodegradable organic P | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A phosphorus containing particulate organic material that is not biologically or chemically degraded in the plant and leaves with the cake solids. The only process that degrades XP,U to SPO4 is the process model of thermal hydrolysis process. | g P.m-3 |
| SO2 | Dissolved oxygen (O2) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Dissolved oxygen is major electron acceptor for aerobic system and is subjected to gas transfer. | g O2.m-3 |
| SCH4 | Dissolved methane (CH4) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Dissolved methane is the most reduced form of carbon and end product of methanogensis. They are subjected to gas transfer. | g COD.m-3 |
| SH2 | Dissolved hydrogen (H2) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Dissolved hydrogen is generated during fermentation and transformed to SCH4 during methanogensis. They are subjected to gas transfer. | g COD.m-3 |
| SALK | Alkalinity (ALK) | MiniSumo | Assumed to be bicarbonate and used for appropriate conversion of electric charges in the biokinetic reactions. | eq ALK.L-1 |
| SCO2 | Total inorganic carbon (CO2) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A sum of carbonic acid, bicarbonate ion, carbonate ion. It is end product of many biological reactions. As bicarbonate it is a substrate for nitrification and also consumed during methanogenesis. | g TIC.m-3 |
| XINORG | Inorganics in influent and biomass | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | It represents the inorganic inerts present in influent and the biomass (0.11 g TSS.g COD-1). It has a SNa, SCl, SCa and SMg content. | g TSS.m-3 |
| SCAT | Other strong cations (as Na+) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Represented as sodium and is an essential element for biomass. It participates in pH and precipitation reactions. | g Na.m-3 |
| SAN | Other strong anions (as Cl-) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Represented as chloride and is an essential element for biomass. It participates in pH and precipitation reactions. | g Cl.m-3 |
| SCa | Calcium | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | An essential element for biomass and XPP and participates in pH and precipitation reactions. | g Ca.m-3 |
| SMg | Magnesium | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | An essential element for biomass and XPP. It participates in pH and precipitation reactions. | g Mg.m-3 |
| SK | Potassium | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | An essential element for biomass and XPP. It participates in pH and precipitation reactions. | g K.m-3 |
| SH2S | Hydrogen sulfide (H2S) | Sumo2S | Dissolved hydrogen sulfide gas, its subjected to gas transfer, biological and chemical reactions. | g S.m-3 |
| SSO4 | Sulfate (SO4) | Sumo2S | An end product of SH2S oxidation. It participates in pH determination | g S.m-3 |
| XS | Particulate elemental sulfur (S) | Sumo2S | An intermediary product of partial SH2S oxidation. | g S.m-3 |
| XFeOH | Ferric hydroxide compounds (FeOH) | MiniSumo | Phosphorus-binding capacity of ferric hydroxides | g Fe.m-3 |
| XFeP | Ferric phosphate compounds (FeP) | MiniSumo | This component results from binding phosphorus to the XFeOH. | g Fe.m-3 |
| XAlOH | Aluminium hydroxide compounds (AlOH) | MiniSumo | It stands for phosphorus-binding capacity of possible Al hydroxides | g Al.m-3 |
| XAlP | Aluminium phosphate compounds (AlP) | MiniSumo | This component results from binding phosphorus to the XAlOH. | g Al.m-3 |
| SFe2 | Ferrous ion (Fe2) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Ferrous ion oxidized to ferric ion and then participates in XHFO formation. It precipiates SH2S to XFeS and SPO4 to XVivi under anaerobic environments. | g Fe.m-3 |
| XHFO,H | Active hydrous ferric oxide, high surface (HFO,H) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A ferric hydroxide with high surface area and high capacity to bind SPO4. Higher mixing condition results in more XHFO,H formation compared to XHFO,L. | g Fe.m-3 |
| XHFO,L | Active hydrous ferric oxide, low surface (HFO,L) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A ferric hydroxide with low surface area and low capacity to bind SPO4. Poor mixing condition results in more XHFO,L formation compared to XHFO,H. | g Fe.m-3 |
| XHFO,old | Aged unused hydrous ferric oxide (HFO,old) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | When there is excess ferric addition and not much SPO4 is available then XHFO,H and XHFO,L formed age into XHFO,old. These are incapable of removing SPO4. | g Fe.m-3 |
| XHFO,H,P | P-bound hydrous ferric oxide, high surface (HFO,H,P) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | The Fe-SPO4 complex formed with XHFO,H. | g Fe.m-3 |
| XHFO,L,P | P-bound hydrous ferric oxide, low surface (HFO,L,P) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | The Fe-SPO4 complex formed with XHFO,L. | g Fe.m-3 |
| XHFO,H,P,old | Aged P-bound hydrous ferric oxide, high surface (HFO,H,P,old) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Aged form of XHFO,H,P. | g Fe.m-3 |
| XHFO,L,P,old | Aged P-bound hydrous ferric oxide, low surface (HFO,L,P,old) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Aged form of XHFO,L,P. | g Fe.m-3 |
| XHAO,H | Active hydrous aluminium oxide, high surface (HAO,H) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Aluminium hydroxide with high surface area and high capacity to bind SPO4. Higher mixing condition results in more XHAO,H formation compared to XHAO,L. | g Al.m-3 |
| XHAO,L | Active hydrous aluminium oxide, low surface (HAO,L) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Aluminium hydroxide with low surface area and low capacity to bind SPO4. Poor mixing condition results in more XHAO,L formation compared to XHAO,H. | g Al.m-3 |
| XHAO,old | Aged unused hydrous aluminium oxide (HAO,old) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | When there is excess XHAO and not much SPO4 is available then XHAO and XHAO,L formed age into XHAO,old. These are incapable of removing SPO4. | g Al.m-3 |
| XHAO,H,P | P-bound hydrous aluminium oxide, high surface (HAO,H,P) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | The Al-SPO4 complex formed with XHAO,H. | g Al.m-3 |
| XHAO,L,P | P-bound hydrous aluminium oxide, low surface (HAO,L,P) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | The Al-SPO4 complex formed with XHAO,L. | g Al.m-3 |
| XHAO,H,P,old | Aged P-bound hydrous aluminium oxide, high surface (HAO,H,P,old) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Aged form of XHAO,H,P. | g Al.m-3 |
| XHAO,L,P,old | Aged P-bound hydrous aluminium oxide, low surface (HAO,L,P,old) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Aged form of XHAO,L,P. | g Al.m-3 |
| XCaCO3 | Calcium carbonate (CaCO3) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Essential chemical for pH control - a precipitate that forms readily | g TSS.m-3 |
| XACP | Amorphous calcium phosphate (ACP) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Elemental composition of Ca3(PO4)2 * 4H2O. | g TSS.m-3 |
| XBSH | Brushite (BSH) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Elemental composition of CaHPO4 * 2H2O. Forms at lower pH values than Struvite | g TSS.m-3 |
| XSTR | Struvite (STR) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Formed under anaerobic environments and has an elemental composition of MgNH4PO4 * 6H2O. | g TSS.m-3 |
| XVivi | Vivianite (Vivi) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Formed under anaerobic environments and has an elemental composition of Fe3(PO4)2 * 8H2O. | g TSS.m-3 |
| XFeS | Iron sulfide (FeS) | Sumo2S | Formed under anaerobic environments and is a precursor for pyrite formation. | g TSS.m-3 |
| H | Enthalpy | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | An energy-like property that is used to calculate temperature of a system. It flows from the influent unit to the plant. | MJ.m-3 |
| SALPHA | Alpha indicator | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A surrogate representing the surfactant composition in the wastewater. | unitless |
| SORPswitch | ORP driver for CASTO activity switches | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | A surrogate switch to calculate the activity of CASTOs. | unitless |
| GCO2 | Carbon dioxide gas (CO2) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Gaseous form represented in bubbles of wastewater system. | g TIC.m-3 |
| GCH4 | Methane gas (CH4) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Gaseous form represented in bubbles of wastewater system. | g COD.m-3 |
| GH2 | Hydrogen gas (H2) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Gaseous form represented in bubbles of wastewater system. | g COD.m-3 |
| GO2 | Oxygen gas (O2) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Gaseous form represented in bubbles of wastewater system. | g O2.m-3 |
| GNH3 | Ammonia gas (NH3) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Gaseous form represented in bubbles of wastewater system. | g N.m-3 |
| GN2 | Nitrogen gas (N2) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Gaseous form represented in bubbles of wastewater system. | g N.m-3 |
| GNO | Nitric oxide gas (NO) | Sumo4N | Gaseous form represented in bubbles of wastewater system. | g N.m-3 |
| GN2O | Nitrous oxide gas (N2O) | Sumo4N | Gaseous form represented in bubbles of wastewater system. | g N.m-3 |
| GH2S | Hydrogen sulfide gas (H2S) | Sumo2S | Gaseous form represented in bubbles of wastewater system. | g S.m-3 |
| GCO2,atm | Carbon dioxide gas (CO2) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Gaseous form represented in the atmosphere around wastewater systems. | %v/v |
| GCH4,atm | Methane gas (CH4) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Gaseous form represented in the atmosphere around wastewater systems. | %v/v |
| GH2,atm | Hydrogen gas (H2) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Gaseous form represented in the atmosphere around wastewater systems. | %v/v |
| GO2,atm | Oxygen gas (O2) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Gaseous form represented in the atmosphere around wastewater systems. | %v/v |
| GNH3,atm | Ammonia gas (NH3) | Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Gaseous form represented in the atmosphere around wastewater systems. | %v/v |
| GN2,atm | Nitrogen gas (N2) | Mini_Sumo, Sumo1, Sumo2, Sumo2C, Sumo2S, Sumo4N | Gaseous form represented in the atmosphere around wastewater systems. | %v/v |
| GNO,atm | Nitric oxide gas (NO) | Sumo4N | Gaseous form represented in the atmosphere around wastewater systems. | %v/v |
| GN2O,atm | Nitrous oxide gas (N2O) | Sumo4N | Gaseous form represented in the atmosphere around wastewater systems. | %v/v |
| GH2S,atm | Hydrogen sulfide gas (H2S) | Sumo2S | Gaseous form represented in the atmosphere around wastewater systems. | %v/v |
| SMM,index | Methyl mercaptan production index | Sumo2S | A surrogate for indicating the potential for odor generation at a wastewater facility. | Unitless |
¶ Biological processes
¶ BOD removal
BOD removal from is characterized as soluble substrate component uptake by biomass and used for growth and respiration.
The colloidal biodegradable components are flocculated into particulate components (see Flocculation paragraph). The particulate components are hydrolyzed into soluble components thus biomass can grow on them (see Hydrolysis paragraph).
¶ Biomasses involved in BOD removal
¶ Ordinary heterotrophic organisms, XOHO
Generalist facultative ordinary hetrotrophic organisms that consume different soluble biodegradable organics including SB, SVFA, and XMEOL and can perform biological removal under aerobic, anoxic, and anaerobic environments. They are also responsible for hydrolysis of the particulates. In MiniSumo and Sumo1, they perform one step denitrification, meaning from SNOx to SN2. In other models they follow two steps, SNO3 reduction to SNO2, and SNO2 reduction to SN2.
| Biological processes | Concepts description |
|---|---|
| Aerobic growth on VFA, O2 |
Growth on SVFA under aerobic conditions. Requires SO2, nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH. |
| Anoxic growth on VFA, NO2- |
Growth on SVFA under anoxic conditions. This is the second step of denitrification in Sumo2, Sumo2C and Sumo2S), SNO2 is reduced into SN2. This process is not described in Sumo1 and MiniSumo. Requires SNO2, nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH. |
| Anoxic growth on VFA, NO3- |
Growth on SVFA under anoxic conditions. This is the denitrification process in Sumo1 and MiniSumo: SNO3 is reduced in SN2. This is the first step of denitrification in Sumo2, Sumo2C and Sumo2S): SNO3 is reduced into SNO2. Requires SNO3, nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH. |
| Aerobic growth on SB, O2 |
Growth on SB under aerobic conditions. Requires SO2, nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH |
| Anoxic growth on SB, NO2- |
Growth on SB under anoxic conditions. This is the second step of denitrification in Sumo2, Sumo2C and Sumo2S), SNO2 is reduced into SN2. This process is not described in Sumo1 and MiniSumo. Requires SNO2, nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH |
| Anoxic growth on SB, NO3- |
Growth on SB under anoxic conditions. This is the denitrification process in Sumo1 and MiniSumo: SNO3 is reduced in SN2. This is the first step of denitrification in Sumo2, Sumo2C and Sumo2S): SNO3 is reduced into SNO2. Requires SNO3, nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH. |
| SB fermentation with high VFA (OHO growth, anaerobic) |
Growth on SB under anaerobic conditions. OHO Fermenters have different growth rate of 0.3 d-1 and digesters have different KSB,ana of 350 mgCOD/L Produce SVFA as fermentation product and SH2. Under high SVFA concentration, the yield of SH2 production is higher (less SVFA produced). Requires nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH. |
| SB fermentation with low VFA (OHO growth, anaerobic) |
Growth on SB under anaerobic conditions. OHO Fermenters have different growth rate of 0.3 d-1 and digesters have different KSB,ana of 350 mgCOD/L Produce SVFA as fermentation product and SH2. Under low SVFA concentration, the yield of SH2 production is lower (more SVFA produced). Requires nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH. |
| Aerobic growth on SMEOL, O2 |
OHO aerobic growth on methanol in order to consum any residual methanol from anoxic carbon dosage for denitrification. Requires SO2, nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH. |
| OHO decay | OHO decay process under anoxic and aerobic conditions. This process release XB,e and XE (death-regeneration concept). |
| OHO anaerobic decay | OHO decay process under anaerobic conditions. This process release XB,e and XE,ana (death-regeneration concept). |
¶ Anoxic methanol utilizers, XMEOLO
Specialist heterotrophic organisms responsible for removal of XMEOL. Only found in systems with methanol addition and compete with OHOs during denitrification process. In Sumo1, they perform one step denitrification, meaning from SNOx to SN2. Is other models they follow two steps, SNO3 reduction to SNO2, and SNO2 reduction to SN2.
| Biological processes | Concepts description |
|---|---|
| MEOLO growth, NO2 |
Growth on SMEOL under anoxic conditions. This is the second step of denitrification in Sumo2, Sumo2C and Sumo2S), SNO2 is reduced into SN2. This process is not described in Sumo1 and MiniSumo. Requires SNO2, nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH |
| MEOLO growth, NO3 |
Growth on SMEOL under anoxic conditions. This is the denitrification process in Sumo1 and MiniSumo: SNO3 is reduced in SN2. This is the first step of denitrification in Sumo2, Sumo2C and Sumo2S): SNO3 is reduced into SNO2. Requires SNO3, nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH. |
| MEOLO decay | MEOLO decay process under anoxic and aerobic conditions. This process release XB,e and XE (death-regeneration concept). |
| MEOLO anaerobic decay | MEOLO decay process under anaerobic conditions. This process release XB,e and XE,ana (death-regeneration concept). |
¶ Nitrogen removal
¶ Nitrification model
Nitrification rate is the most important parameter in design and reliable simulation of BNR (Biological Nutrient Removal) plants. The maximum specific growth rate of nitrifiers ranges from 0.2 to 1 d-1 for different wastewater. This parameter should be considered as part of wastewater characterization.
There are three models available in Sumo to describe the process on three layer of details:
- Mini_Sumo and Sumo1 – one step nitrification process
- one nitrifying organism (XNITO),
- ammonia (SNHx) and nitrate (SNOx) as nutrients
- Sumo2, Sumo2C, and Sumo2S – two step nitrification process
- Three organisms
- ammonia oxidizing bacteria (XAOB)
- nitrite oxidizing bacteria (XNOB)
- Anamox organisms (XAMX)
- Three nutrients
- Ammonia (SNHx), nitrite (SNO2), nitrate (SNO3)
- XAOB kinetics are the first step and rate limiting
- Three organisms
- Sumo4N - detailed nitrification and GHG emission model
- 4-step nitrification model from Pocquet et al., 2016
- 4-step denitrification model from Hiatt and Grady, 2008
- NO considered as cell internal intermediate
Sumo4N model is describe in details in Focus models mechanisms chapter.
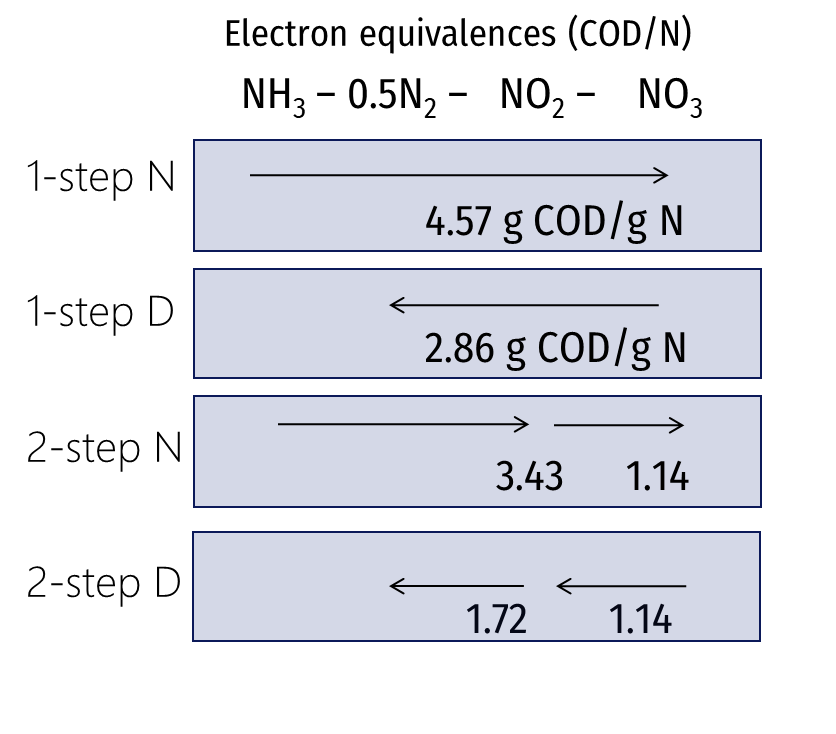
¶ Biomasses involved in nitrification
¶ Aerobic nitrifying organisms, XNITO
Obligate aerobic autotrophic organism responsible for complete nitrification, from SNHx to SNOx in MiniSumo and Sumo1. It represents a combination of AOBs and NOBs for simplification in the referred models.
| Biological processes | Concepts description |
|---|---|
| NITO growth |
NITO growth process, using SNHx as electron donor. SNHx is oxidized in SNO3 in one step. SCO2 is used as carbon source. Requires SO2, nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH. |
| NITO decay | NITO decay process under anoxic and aerobic conditions. This process release XB,e and XE. |
| NITO anaerobic decay | NITO decay process under anaerobic conditions. This process release XB,e and XE,ana . |
¶ Aerobic ammonia oxidizers, XAOB
Obligate aerobic autotrophic organism responsible for the first step in nitrification, from SNHx to SNO2 in Sumo2, Sumo2C and Sumo2S.
| Biological processes | Concepts description |
|---|---|
| AOB growth |
AOB growth process, using SNHx as electron donor. This is the first step of nitrification: SNHx is oxidized in SNO2 SCO2 is used as carbon source. Requires SO2, nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH. |
| AOB decay | AOB decay process under anoxic and aerobic conditions. This process release XB,e and XE. |
| AOB anaerobic decay | AOB decay process under anaerobic conditions. This process release XB,e and XE,ana . |
¶ Nitrite oxidizers, XNOB
Obligate aerobic autotrophic organism responsible for the second step in nitrification, from SNO2 to SNO3 in Sumo2, Sumo2C and Sumo2S.
| Biological processes | Concepts description |
|---|---|
| NOB growth |
NOB growth process, using SNO2 as electron donor. This is the second step of nitrification: SNO2 is oxidized in SNO3 SCO2 is used as carbon source. Requires SO2, nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH. |
| NOB decay | NOB decay process under anoxic and aerobic conditions. This process release XB,e and XE. |
| NOB anaerobic decay | NOB decay process under anaerobic conditions. This process release XB,e and XE,ana . |
¶ Anammox organisms, XAMX
Anammox is a chemoautotrophic anaerobic bacteria that oxidizes ammonium with nitrite as the electron acceptor and with CO2 as the main carbon source. They are implemented in Sumo2, Sumo2C, Sumo2S and Sumo4N models.
Anammox are slow growers and have a long doubling time. Their growth reaction stoichiometry is described by Strous (1998, 1999):
NH4+ + 1.32 NO2- + 0.066 HCO3- + 0.13 H+ = 0.26 NO3- + 1.02 N2 + 0.066 CH2O0.5N0.15 + 2.03 H2O
This reaction allows saving up to 63% of oxygen demand and 100% of carbon requirement (Figure 2.2) compared to a full nitrification and denitrification pathway for nitrogen removal (Figure 2.1).
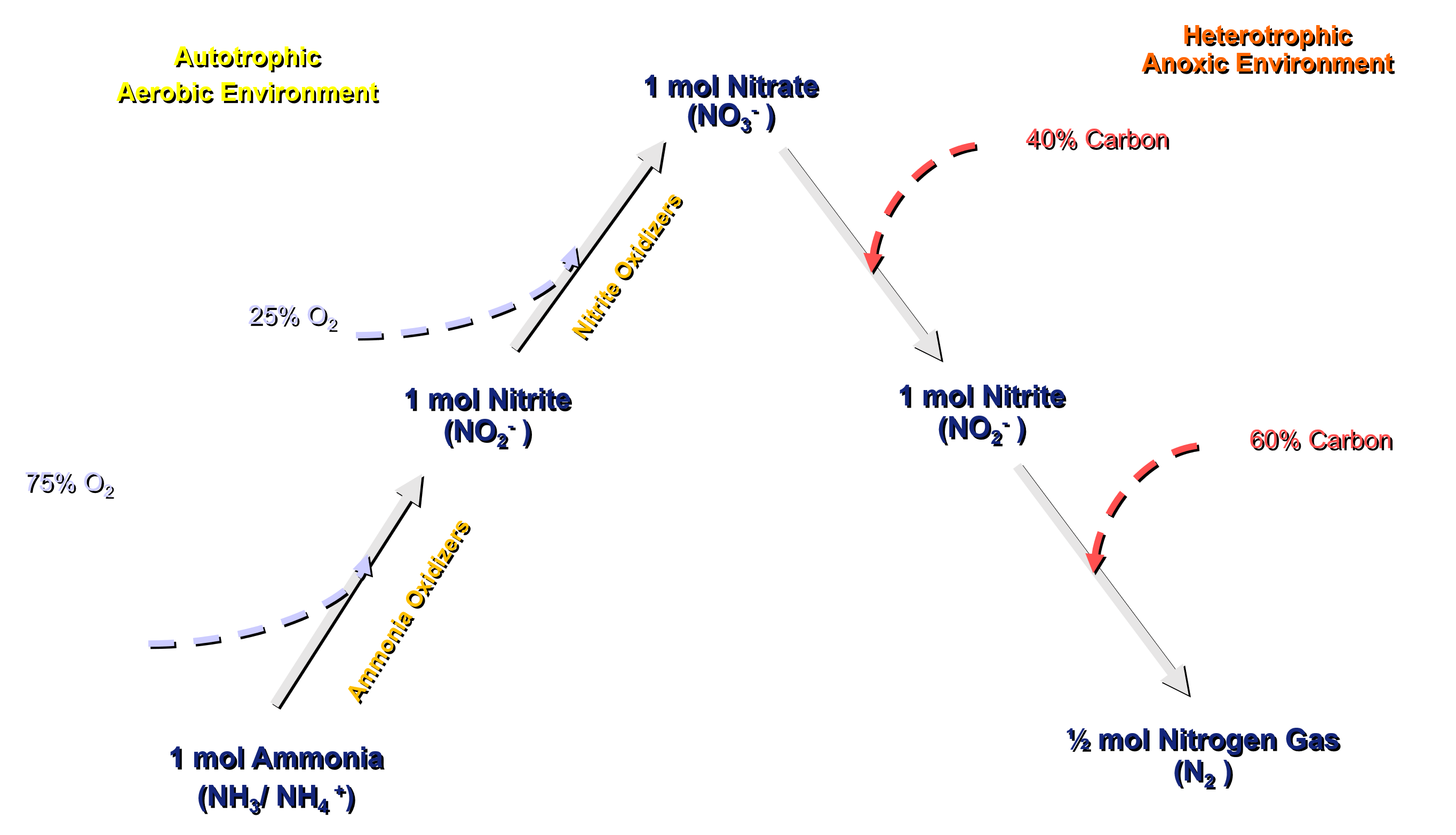
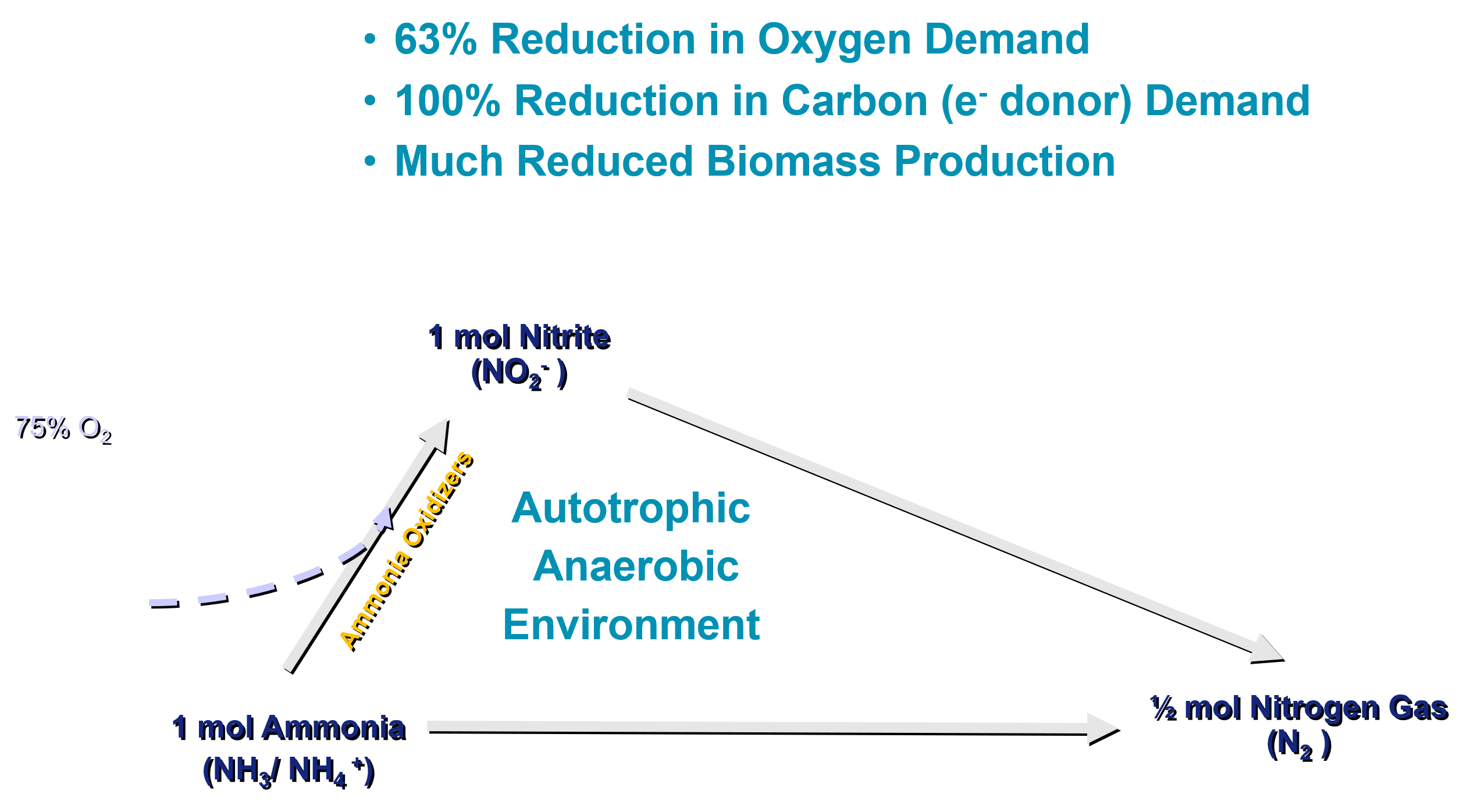
The Anammox organisms are modelled in Sumo2, Sumo2C, Sumo2S and Sumo4N models. The model includes description of anammox growth, decay and anaerobic decay.
The stoichiometry of Anammox growth in Sumo is derived from the stoichiometry proposed by Strous. Two stoichiometric parameters are introduced: the amount of nitrite utilised per mol of ammonia oxidized, and the amount of nitrate produced per mol of ammonia utilised.
| YAMX,NO2 | Yield of AMX on NO2 | 1.32 | mol NO2/mol NH4 |
| YAMX,NO3 | Yield of AMX on NO3 | 0.26 | mol NO3/mol NH4 |
The stoichiometric coefficient expressions are then obtained by applying the full elemental balance methodology from Takacs et al. (2007).
A generic biomass composition CaCHaHOaONaN is considered. The theoretical oxygen demand of this biomass is by definition:
BioThOD = (aC + aH/4 - 3 * aN/4 - aO/2) * 2 * AMO
with AMO as the atomic mass of oxygen.
Considering uncharged species, the Anammox reaction is set up as:
(vNH3 + aN) * NH3 + vHNO2 * HNO2 + aC * CO2 = CaCHaHOaONaN + vN2 * N2 + vHNO3 * HNO3 + vH2O * H2O
With vNH3, vHNO2, vN2, vHNO3 and vH2O the stoichiometric coefficients of the ammonia oxidation with nitrite reaction.
Five equations can be derived from this reaction:
- Two equations for the definitions of the stoichiometric parameters that are chosen:
- YNO2 = vHNO2 /vNH3
- YNO3 =v HNO3 /vNH3
- Three mass balances can be expressed for hydrogen, nitrogen and oxygen:
- H balance: 3 * (vNH3 + aN) + vHNO2 = aH + vHNO3 + 2 * vH2O
- N balance: vNH3 + vHNO2 = vHNO3 + 2 * vN2
- O balance: 2 * vHNO2 + 2 * aC = aO + 3 * vHNO3 + vH2O
These 5 equations can be used to find the 5 unknowns: vNH3, vHNO2, vHNO3, vN2 and vH2O.The following code is executed in python for the symbolic resolution:
from sympy import symbols, solve, Eq
NH3, vNH3, HNO2, vHNO2, CO2, H3PO4, N2, vN2, HNO3, vHNO3, H2O, vH2O, YNO2, YNO3, aC, aH, aO, aN =
symbols('NH3, vNH3, HNO2, vHNO2, CO2, H3PO4, N2, vN2, HNO3, vHNO3, H2O, vH2O, YNO2, YNO3, aC, aH, aO, aN')
solve(( Eq(YNO2, vHNO2*N/(vNH3*N)), Eq(YNO3, vHNO3*N/(vNH3*N)), Eq( 3*(vNH3+aN)+vHNO2,aH+vHNO3+2*vH2O), Eq( vNH3+vHNO2,vHNO3+2*vN2), Eq(2*vHNO2+2*aC,aO+3*vHNO3+vH2O)), vNH3, vHNO2, vHNO3, vN2, vH2O)
The result is (vH2O is not shown as not included in the model):
vNH3 = (4 * aC + aH - 3 * aN - 2 * aO) /(-3 * YNO2 + 5 * YNO3 + 3)
vHNO2 = YNO2 * (4 * aC + aH - 3 * aN - 2 * aO)/(-3 * YNO2 + 5 * YNO3 + 3)
vHNO3 = YNO3 * (4 * aC + aH - 3 * aN - 2 * aO)/(-3 * YNO2 + 5 * YNO3 + 3)
vN2 = (4 * YNO2 * aC + YNO2 * aH - 3 * YNO2 * aN - 2 * YNO2 * aO - 4 * YNO3 * aC - YNO3 * aH + 3 * YNO3 * aN +
+ 2 * YNO3 * aO + 4 * aC + aH - 3 * aN - 2 * aO) /(2 * (-3 * YNO2 + 5 * YNO3 + 3))
These coefficients must be converted per gram of biomass COD (arbitrary reference in all biomass growth reactions) and in g N. Then, vNH3, vHNO2 and vHNO3 are multiplied by AMN/BioThOD and vN2 is multiplied by 2*AMN/BioThOD. The coefficient aN is the nitrogen content of the biomass, iN,Bio. As nitrogen content of the biomass is explicitly expressed, the definition of the theoretical COD of the biomass is revised as:
BioThOD=(aC+aH/4-aO/2)*2*AMO.
The convention of the negative signs for consumed components is also added. This results in the following stoichiometric coefficient expressions:
vNH3 = -(3 * iN,BIO * AMO + 2 * AMN) /(5 * YAMX,NO3 + 3 - 3 * YAMX,NO2) /AMO
vHNO2= -YAMX,NO2 /(5 * YAMX,NO3 + 3 - 3 * YAMX,NO2) * (3 * iN,BIO * AMO + 2 * AMN) /AMO
vHNO3= YAMX,NO3 /(5 * YAMX,NO3 + 3 - 3 * YAMX,NO2) * (3 * iN,BIO * AMO + 2 * AMN) /AMO
vN2 = 2 * (YAMX,NO3 * AMN - 1 * AMN - 1 * YAMX,NO2 * AMN + 4 * YAMX,NO3 * iN,BIO * AMO -
-3 * YAMX,NO2 * iN,BIO * AMO) /AMO /(3 * YAMX,NO2 - 5 * YAMX,NO3 - 3)
| Biological processes | Concepts description |
|---|---|
| Growth |
AMX growth process, using SNHx as electron donor and SNO2 as electron acceptor. SNO3 and SN2 are produced by the reaction. The stoichiometry is described above. SCO2 is used as carbon source. Requires SNHx, SNO2, nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH. |
| Decay | AMX decay process under anoxic and aerobic conditions. This process release XB,e and XE. |
| Anaerobic decay | AMX decay process under anaerobic conditions. This process release XB,e and XE,ana . |
Strous, M., Heijnen, J., Kuenen, J. et al. The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms. Appl Microbiol Biotechnol 50, 589–596 (1998). https://doi.org/10.1007/s002530051340
Strous, M., Fuerst, J., Kramer, E. et al. Missing lithotroph identified as new planctomycete. Nature 400, 446–449 (1999). https://doi.org/10.1038/22749
Takács, I., Vanrolleghem, P.A., Wett, B., Murthy, S., 2007. Elemental balance based methodology to establish reaction stoichiometry in environmental modeling. Water Sci. Technol. 56, 37–41.
¶ Impact of operating conditions and design on nitrification
Nitrification is impacted by:
- dissolved oxygen level
- liquid temperature
- pH and alkalinity
- Low mixing condition or floc size can cause diffusion issues and cause reduction in nitrification rate. Half saturation for oxygen for nitrifiers can be possible calibration factor to mimic this limitation.
If the ammonia level in the aerobic cell is close to half saturation, then its important to simulate the reactor as plug flow.
¶ Sidestream and mainstream nitrification
Two types of process unit model options are available for selection on configure tab: Mainstream or Sidestream (these can be selected from the configure tab if i.e. the CSTR is selected)
The growth rate and half-saturation for sidestream reactors are different due to possible diffusion issues, inhibitory compounds in centrate, and temperature.
¶ Measuring specific growth rate
Two different tests can be performed to measure nitrifiers growth rate:
- High F/M test – 7 days
- Batch test: Small initial nitrifier concentration combined with high initial ammonia concentration
- Exponential response of NOx to estimate growth rate
- pH and alkalinity control are especially important
- This can be done with different wastewater to identify possible inhibition, download and see the High FM.msumo example
- Washout test – several days
- Operating a flow through CSTR just at correct HRT initially filled with nitrifying sludge
- Important step to identify flow rate to the CSTR just enough to cause a washout
- There is little to no growth of nitrifiers during the test and test is performed using actual plant influent
- See Washout test.msumo example
¶ Denitrification model
Denitrification is the reduction process of oxidized forms of oxygen, used as electron acceptor in the growth process of heterotrophic organisms. If denitrification is complete, these electron acceptors are reduced sequentially to nitrogen gas (SN2).
¶ Impact of operating conditions and design on denitrification
Denitrification is impacted by the availability of a carbon source. Knowing RBCOD concentration is extremely important for nitrogen removal design and modelling. The denitrification rate is then controlled by:
- the concentration of readily biodegradable COD (SB plus SVFA) in raw wastewater
- the addition of an external carbon source
- the rate of hydrolysis under anoxic condition (when other carbon sources are depleted)
¶ Model specifications under anoxic conditions
- MiniSumo
- One biomass involved in denitrification: OHOs
- One step denitrification, SNO3 to SN2
- Growth of heterotrophic biomass on SB reduced by reduction factor as nor SVFA and neither methanol utilizers are part of the model
- Decay of OHO also reduced under anoxic condition
- Hydrolysis reduced under denitrification using the reduction factor
- Sumo1
- 3 biomasses involved in denitrification: OHOs, CASTOs and MEOLOs
- One step denitrification, SNO3 to SN2
- Hydrolysis reduced under anoxic conditions
- Growth of OHO on SB and SVFA is reduced under anoxic conditions
- First substrate preference is SVFA over SB, reaction is reduced for SB until SVFA disappears
- Methanol utilizers and their respective kinetics and stoichiometry is included
- PAOs and GAOs perform denitrification on stored compounds and have a reduction factor
- All biomass has reduction factor for decay even for methanol utilizers
- Sumo2, Sumo2C and Sumo2S
- 3 biomasses involved in denitrification: OHOs, CASTOs and MEOLOs
- Two step denitrification: SNO3 to SNO2 to SN2
- All the reactions are same as Sumo1 but with 2 steps of denitrification
- Carbon, nitrate and nitrite switching functions allow to simulate nitrite accumulation in PdNA processes. See PdNA model description below.
- Sumo2S - Sulphur-oxidizing organisms (SOO) are involved in denitrification in presence of H2S and elemental sulphur
- Sumo4N
- Sumo4N model is describe in details in Focus models mechanisms chapter.
¶ PdNA model
Successful implementation of mainstream partial denitrification and anammox (PdNA) process is critical for meeting the future’s stringent effluent total nitrogen targets with low energy demand and carbon addition costs. PdNA relies on dosing enough carbon (of a specific type) to an anoxic zone so that the heterotrophic denitrification produces nitrite (through partial denitrification), which is necessary for the growth of anammox. A good design and identification of an optimized operational strategy is impossible without being able to model such a complex biological process.
The current models do not allow nitrite accumulation based on the type of carbon and the carbon-to-nitrate ratio of the system. This resulted in an inability to evaluate the correct design conditions for growing anammox alongside the heterotrophic denitrifiers, frequently leading to overestimating the carbon dosage.
To support the efforts of design and research engineers, Sumo24 two-step nitrification-denitrification models have been improved to simulate the PdNA process. Based on the carbon-to-nitrate ratio, a mechanism associated with using methanol and volatile fatty acids as the two carbon sources is introduced, which allows for nitrite accumulation under low carbon addition. The model developed is calibrated using data from the literature.
The model behave as follow:
- While there is some NO3, nitrate is denitrified into nitrite under 2 alternative conditions:
- if the VFA is low
- if the VFA is high but there is no NO2
- Denitrification of nitrite to N2 when nitrate is depleted
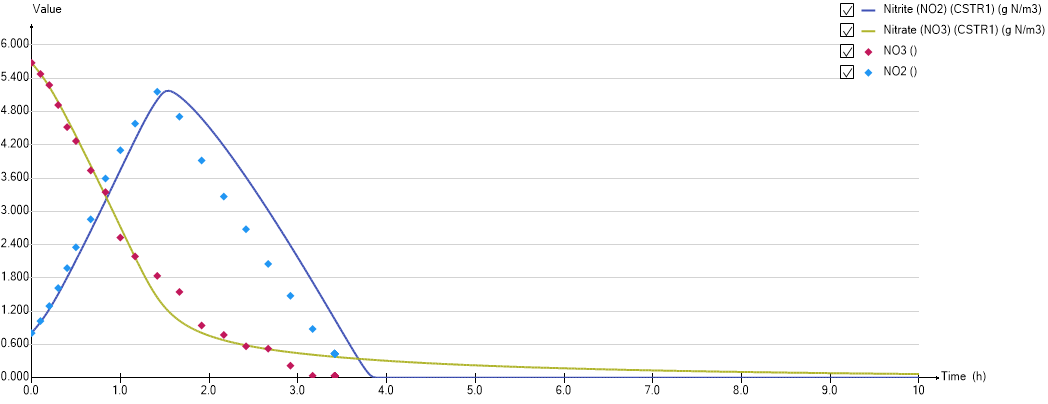
¶
Key model details of the denitrification process
- Under anoxic conditions, the yield coefficients are reduced (compared to aerobic conditions) and is particularly important for denitrification kinetics analysis.
- Reduction factor for anoxic growth kinetic of OHOs
- In Sumo the default value of 0.6 is used for denitrification while
- In real plants it ranges from 0.3 to 0.85 and
- it can depend on system design or operation, represents degree of removal rate and process SRT.
- It can be used for calibration
- Results in lower rate of substrate utilization and endogenous decay. This is because not all the biomass are facultative and don’t participate in denitrification.
- is implemented as relative energy generated is higher with O2 as electron acceptor than with nitrate.
- The actual value be determined by performing OUR and NUR test, see Anoxic reduction factor.msumo
- Denitrification batch tests are done to identify the denitrification rate kinetics, see Denitrification test MLSS.msumo
- In Sumo the default value of 0.6 is used for denitrification while
- Inhibition due to oxygen is simulated using an inverse Monod also referred as a switching function
- (KO2)/((KO2) + (SO2)), dissolved oxygen inhibits denitrification reactions
- This value is 0.15 g O2/m3, however, this is an important constant that can be used to calibrate denitrification performance, especially in case of large flocs, deep tanks, and not well mixed conditions.
- A high value of the constant means, more denitrification under partially aerobic conditions, can be used for calibration
- Specific denitrification rate (g N/g VSS/h) is not a model parameter but is a calculated variable
¶ Phosphorus removal
Biological phosphorus removal is achieved in activated sludge processes via two main pathways:
- Removal by biomass synthesis
Incorporation into molecules building cell walls, DNA, ATP as 1-2% of these is phosphorus. This process can remove at least 30% of P from influent and transfer it to the sludge. - Enhanced biological P removal
This chapter focuses on this process as the “luxury uptake” of polyphosphate once the conditions of the plant are set properly.
¶ Enhanced biological phosphorus removal (EBPR) processes
Sumo uses a model where two biomass fractions compete for the volatile fatty acids (VFA): one of them storing phosphates (phosphorus accumulating organisms, PAO) and the other one storing glycogen (glycogen accumulating organisms, GAO). These biomass fractions are represented as one unified biomass (carbon storing organisms, CASTO) in the model, however the processes that are specific to PAO or GAO are differentiated when external (ambient conditions) or internal (metabolism) driving forces require. The proportion between GAOs and PAOs is tracked by the amount of stored poly-hydroxy-alkanoates (PHA,GAO and PHA,PAO respectively) relative to all stored carbon (XSTC = XPHA,PAO + XPHA,GAO) in the system.
Note that GAOs are storing both PHA and glycogen as carbon storage, but in Sumo only PHA storage is accounted for as a state variable.
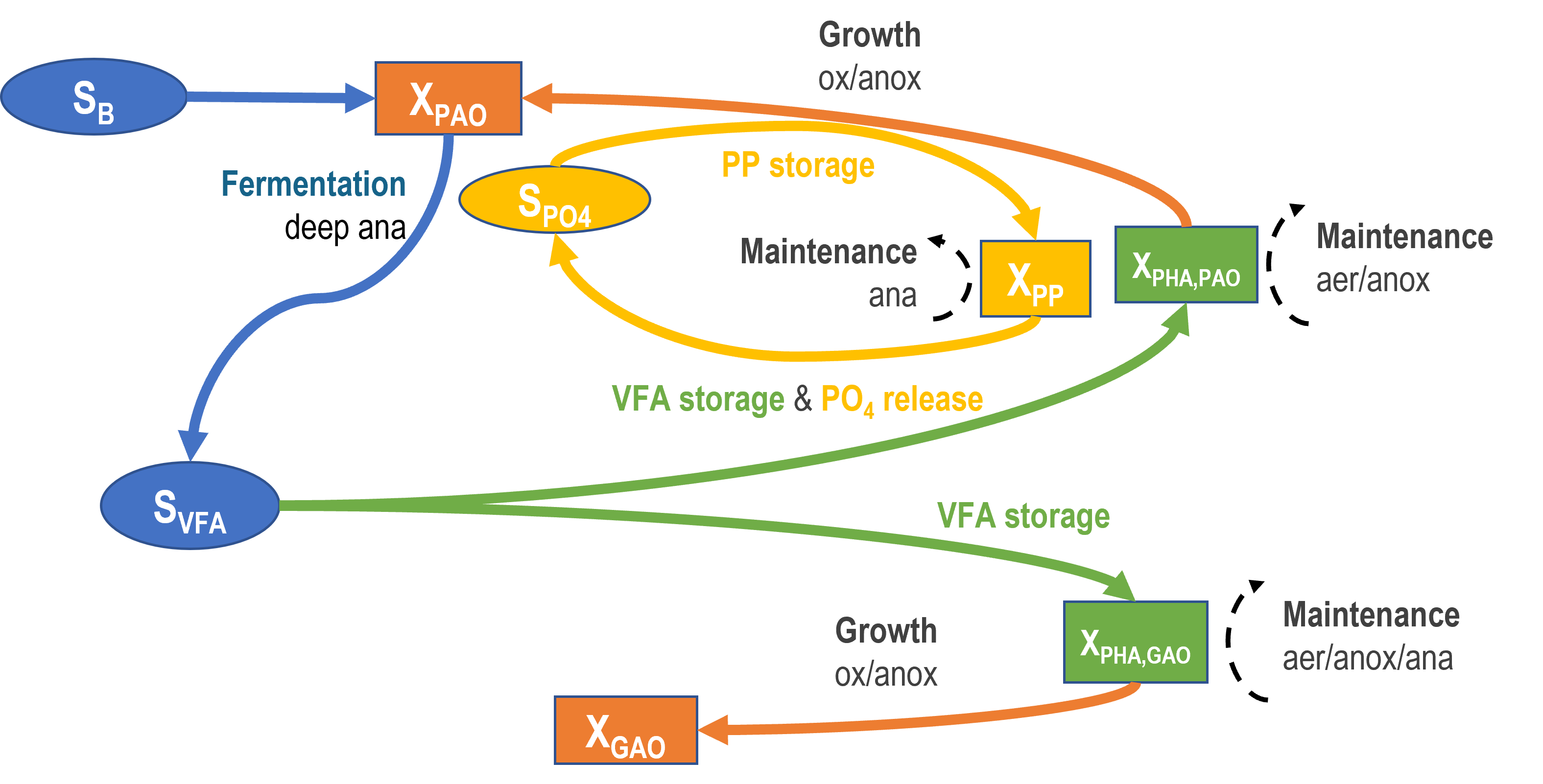
A logistic ORP switch function is employed to control the competition between VFA storage by GAOs and PAOs: at low ORP, GAO storage is inhibited and PAO storage is favored. Another ORP switch function is used to control the fermentation activity of Tetrasphaera-like PAOs under deep ORP regime.
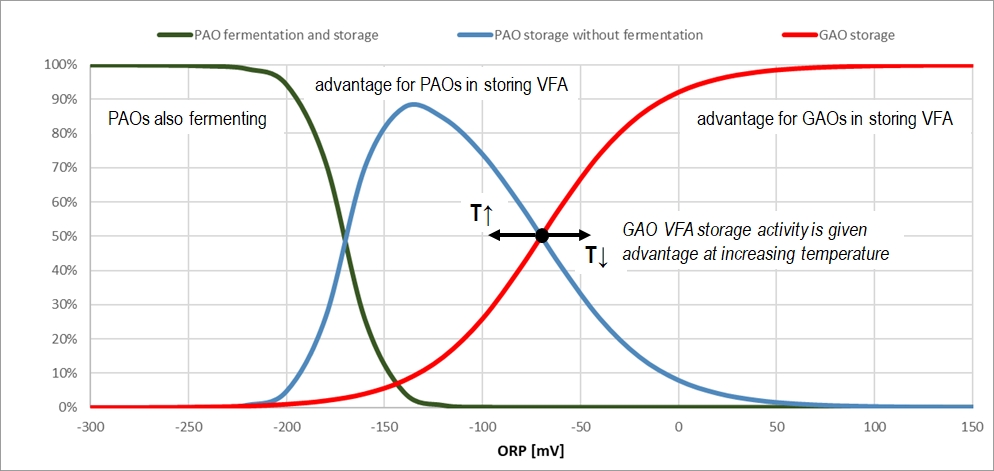
In order to account for the lag in the biomass response following a change in the ORP (which can happen rapidly), a regulating role state variable (SOPRswitch, called as “ORP driver for CASTO activity switches”) was introduced to the model, along with a process that changes the value of this variable according to the following first-order kinetics:
dSORPswitch/dt = kORPswitch * (ORP + offsetORPswitch – SORPswitch)
where:
- ORP is the actual ORP derived from actual concentrations of O2, NOx, CH4 and H2 (as well as H2S in models involving sulfur);
- offsetORPswitch is a simple constant value that transposes SORPswitch values into the positive range (required by the solver algorithm);
- kORPswitch is the kinetic rate that represents the ORP time lag.
The effect is that SORPswitch follows ORP with a lag depending on the actual difference from the actual ORP. For small changes in the latter, SORPswitch is quick to catch up, while for abrupt changes it takes time for SORPswitch to catch up with ORP, resulting in a less hectic timeline of SORPswitch compared to ORP. The response time can be adjusted by manipulating the kORPswitch parameter. The default value of the kORPswitch parameter is 24 d-1, to allow for changes to happen in the order of magnitude of an hour (accommodating the model to be applied to dynamic systems like SBRs using the default parameter).
The PAO/GAO VFA storage mechanisms have been set in all plantwide process models to be driven by the new SORPswitch variable instead of ORP.
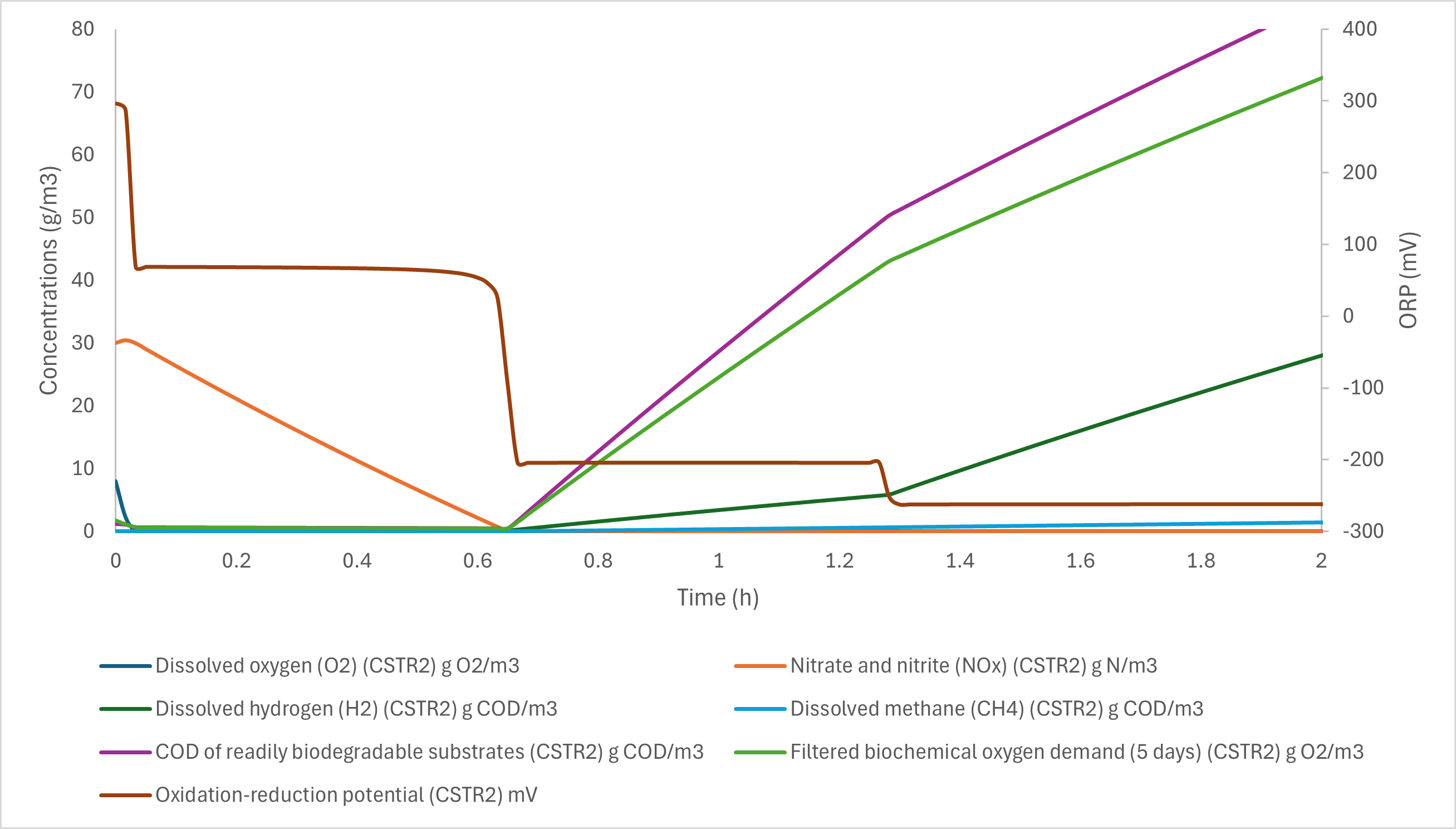
¶ ORP model
The oxidation-reduction potential is usually difficult to measure accurately in wastewater. The ORP value is estimated in the model, depending on the conditions:
- Under aerobic conditions, it considers a maximum of +300 mV value, that is modulated with the depletion of dissolved oxygen towards the anoxic maximum value (+70 mV)
- Under anoxic conditions, it considers a maximum of +70 mV value, that is modulated with the depletion of nitrates plus nitrites
- Under anaerobic conditions, the ratio of the production rate of H2 and CH4 (calculated as Anaerobic Activity Rate AAR), related to the rbCOD degradation rate (SVFA, SB and SMEOL degradation rate) is used as an indicator of the anaerobic activities. The higher this ratio, the lower the ORP value.
The final ORP is the maximum of the aerobic, anoxic and anaerobic ORP.
¶ Biomass involved in BioP: Carbon storing organisms, XCASTO
| Biological processes | Concepts description |
|---|---|
| CASTO growth on PHA,PAO and PHA,GAO, O2 |
PHA,PAO/PHA,GAO → CASTO Kinetic parameters
Kinetic rate limitation/inhibitions
|
| CASTO growth on PHA,PAO and PHA,GAO, NOx/NO2/NO3 |
PHA,PAO/PHA,GAO → CASTO Kinetic parameters
Kinetic rate limitation/inhibitions
|
| PAO polyphosphate storage, O2 |
SPO4 → XPP Kinetic parameters
Kinetic rate limitation/inhibitions
|
| PAO polyphosphate storage, NOx/NO2/NO3 |
SPO4 → XPP Kinetic parameters
Kinetic rate limitation/inhibitions
|
| PAO growth on PHA,PAO, O2; PO4 limited |
PHA,PAO → CASTO Kinetic parameters
Kinetic rate limitation/inhibitions
|
| PAO growth on PHA,PAO, NOx/NO2/NO3; PO4 limited |
PHA,PAO → CASTO Kinetic parameters
Kinetic rate limitation/inhibitions
|
| PAO's PHA,PAO storage from VFAs and PO4 release |
VFA → PHA,PAO and XPP → SPO4 Kinetic parameters
Kinetic rate limitation/inhibitions
|
| GAO's PHA,GAO storage from VFAs |
VFA → PHA,GAO Kinetic parameters
Kinetic rate limitation/inhibitions
|
| CASTO aerobic maintenance |
PHA,PAO → CO2 and PHA,GAO → CO2 Requires O2. Kinetic parameters
Kinetic rate limitation/inhibitions
|
| CASTO anoxic maintenance, NOx/NO2/NO3 |
PHA,PAO → CO2 and PHA,GAO → CO2 Requires NOx/NO2/NO3. Kinetic parameters
Kinetic rate limitation/inhibitions
|
| GAO anaerobic maintenance |
PHA,GAO → VFA Kinetic parameters
Kinetic rate limitation/inhibitions
|
| PP cleavage for maintenance |
PP → PO4 Rate expression covers the aerobic, anoxic and anaerobic PP cleavage as follows (Sumo1, MODEL sheet, row 27, column CB): r24 = bPP,ana,T * XPAO * LogsatXPP,KPO4,PAO * (ηbPP,aer * MRinhXPHA,XPAO,KPHA,cle * MsatSO2,KO2,CASTO + ηbPP,anox * MinhSO2,KO2,CASTO * MsatSNOx,KNOx,CASTO + MinhSO2,KO2,CASTO * MinhSNOx,KNOx,CASTO) Kinetic parameters
Kinetic rate limitation/inhibitions
|
| SB fermentation with high VFA (PAO growth, anaerobic) |
SB → VFA + CASTO Kinetic parameters
Activity limitation
Kinetic rate limitation/inhibitions
|
| SB fermentation with low VFA (PAO growth, anaerobic) |
SB → VFA + CASTO Kinetic parameters
Activity limitation
Kinetic rate limitation/inhibitions
|
| CASTO decay |
CASTO → XB,e and XE under anoxic and aerobic conditions. Kinetic parameters
Kinetic rate limitation/inhibitions
|
| CASTO anaerobic decay |
CASTO → XB,e and XE under anaerobic conditions. Kinetic parameters
Kinetic rate limitation/inhibitions
|
¶ Chemical phosphorus removal
Phosphorus can be chemically removed by:
- metal salt addition (iron or aluminium). The chemical processes implemented in the model are described in a dedicated paragraph)
- natural or induced precipitation of:
- Amorphous calcium phosphate (Ca3(PO4)2 * 4H2O)
- Brushite (CaHPO4 * 2H2O)
- Struvite (MgNH4PO4 * 6H2O)
- Vivianite (Fe3(PO4)2 * 8H2O)
The chemical precipitation model is described in a dedicated paragraph.
¶ Anaerobic digestion processes
All Sumo models describe the anaerobic digestion processes involving 3 biomasses: the OHOs with fermentation process, Acidoclastic methanogens (AMETOs) and hydrogenotrophic methanogens (HMETOs).
The typical pathway of anaerobic digestion is the lysis of all biomasses (except OHOs and methanogens), hydrolysis of biodegradable particulate substrate, fermentation and methanogenesis, leading to methane production.
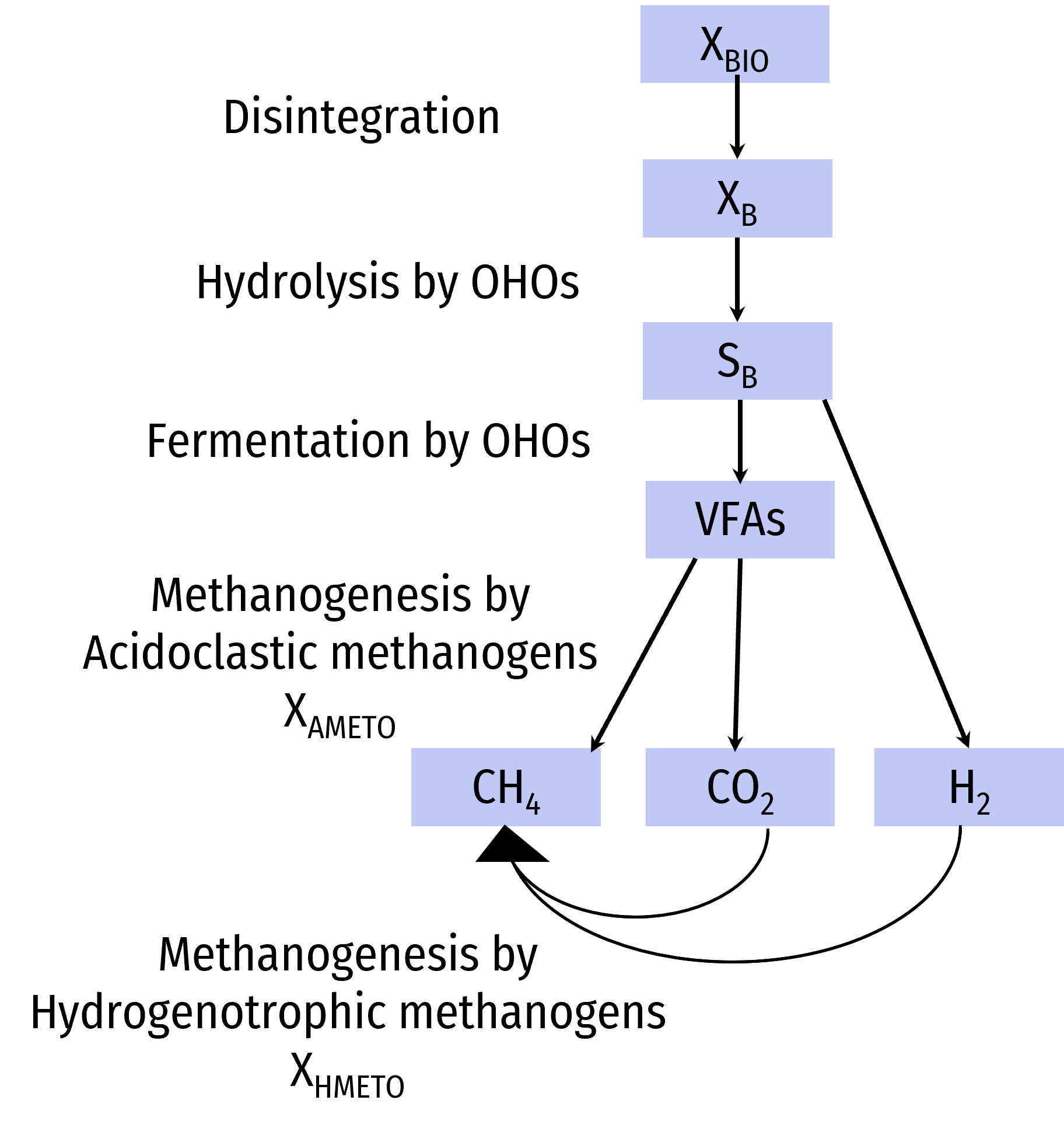
The OHOs fermentation proces is described in BOD removal paragraph.
¶ Methanogenesis
¶ Acidoclastic methanogens, XAMETO
Anaerobic archea that consume SVFA to produce SCH4 and SCO2 as a metabolic by-product and don’t grow under aerobic conditions.
| Biological processes | Concepts description |
|---|---|
| AMETO growth |
Substrate is SVFA and produce SCH4. Too much SVFA results in a Haldane SVFA inhibition Free ammonia inhibition Requires nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH. |
| AMETO decay | AMETO decay process under anoxic and aerobic conditions. This process release XB,e and XE (death-regeneration concept). |
| AMETO anaerobic decay | AMETO decay process under anaerobic conditions. This process release XB,e and XE,ana (death-regeneration concept). |
¶ Hydrogenotrophic methanogens, XHMETO
Anaerobic archea that consume SH2 and SCO2 to produce SCH4 as a metabolic by-product and don’t grow under aerobic conditions.
| Biological processes | Concepts description |
|---|---|
| HMETO growth |
Substrate H2 and produce SCH4. Bell shape inhibition function on pH: BellinhpH. |
| HMETO decay | HMETO decay process under anoxic and aerobic conditions. This process release XB,e and XE (death-regeneration concept). |
| HMETO anaerobic decay | HMETO decay process under anaerobic conditions. This process release XB,e and XE,ana (death-regeneration concept). |
¶ Biological conversions
¶ Flocculation
Flocculation is a process where colloidal components in liquid phase are aggregated into larger flocs thus behave as particulates. In Sumo models flocculation process is a conversion of colloidal components into particulates. The process rate is depending on the concentration of total biomass.
The addition of metal salts (iron as HFO or alum as HAO) increase the flocculation rate of colloidal material, and can be used as a chemical addition for this purpose in primary settlers.
¶ Hydrolysis
Hydrolysis is a process where larger molecules are broken into smaller molecules which are soluble in liquid phase and available for bacterial growth with a certain delay (enzymatic processing, chemical dissolution, mass transport, storage, etc.).
Depending on the origin of the organic molecules, two types of hydrolysis reactions can be distinguished: hydrolysis of ‘‘primary substrate’’ that comes from the influent and hydrolysis of the matter produced by biomass metabolism or decay, named ‘‘secondary substrate,’’ in which protozoa may play an important role (Morgenroth et al., 2002). Consequently, models using the death-regeneration concept to model biomass decay, as Sumo models, merge those two types of hydrolysis in a single process, whereas in case of the endogenous respiration concept, the hydrolysis of secondary substrate is modeled through endogenous respiration and maintenance processes.
Several authors have implemented parallel hydrolysis of particulate organics with different hydrolysis rate (Ohron et al, 1998; Larrea et al., 2002). Anaerobic digestion modeling of different proportions of primary and secondary sludge in the feed have been proved to be better simulated with two particulate substrates: one particulate substrate from influent with higher hydrolysis rate, and one particulate substrate from biomass decay with a lower hydrolysis rate.
In Sumo hydrolysis is a biological process where particulate components are converted into soluble components. Two particulate substrates are distinguished:
- XB coming with the influent (primary substrate)
- XB,e coming from the biomass decay (secondary substrate)
The process rate is higher for XB (3 d-1) than for XB,e (1 d-1) and depend on the concentration of heterotroph biomass.
Larrea, L., Irizar, I., Hildago, M.E., 2002. Improving the predictions of ASM2d through modelling in practice. Water Sci. Technol. 45, 199–208.
Morgenroth, E., Kommedal, R., Harremoes, P., 2002. Processes and modeling of hydrolysis of particulate organic matter in aerobic wastewater treatment - A review. Water Sci. Technol. 45, 25–40.
Orhon, D., Cokgor, E.U., Sozen, S., 1998. Dual hydrolysis model of the slowly biodegradable substrate in activated sludge systems. Biotechnol. Tech. 12, 737–741.
Ozyildiz, G., Zengin, G.E., Güven, D., Cokgor, E., Özdemir, Ö., Hauduc, H., Takács, I., Insel, G., 2023. Restructuring anaerobic hydrolysis kinetics in plant-wide models for accurate prediction of biogas production. Water Res. 245, 120620. https://doi.org/1ce0.1016/j.watres.2023.120620
¶ Ammonification
The ammonification process is part of the nitrogen cycle when organic nitrogen is turned into soluble ammonia. In Sumo models soluble organic nitrogen compound is converted into ammonia. The process rate is depending on the concentration of heterotroph biomass.
¶ Biological phosphorus conversion
Biological phosphorus conversion is similar to the ammonification process but in this process organic phosphorus is converted into phosphate. The process rate is depending on the concentration of heterotroph biomass.
¶ Endogenous products conversion
In Sumo model this process is used to mimic the extreme slow hydrolysis of endogenous decay products by slowly converting XE to particulate biodegradable substrate. The process rate is depending on the concentration of heterotroph biomass.
¶ Anaerobic methanol conversion
Fermentation process of methanol utilizer organisms under anaerobic condition. The process rate depends on the methanol utilizer organisms concentration.
¶ Nitrate/Nitrite assimilative reduction
Processes that occur only under limited ammonia availability. It ensures sufficient ammonia production for biomasses growth by reducing SNO3 and SNO2 into SNHx with the biomasses as electron donors.
¶ Photosynthesis processes
Photosynthetic processes are active only in the Gen3 pond process unit. The photosynthetic organisms (XALGAE) are considered as one biomass group which can grow, respire and decay.
Growth is dependent on the user input "Solar radiation for algal photosynthesis - depth averaged" which may vary depending on time of day (there is no sunlight at night), latitude and season, cloud cover as well as the depth of the pond. It is the user's responsibility to account for these factors when specifying the depth averaged solar radiation. Specifying diurnal and seasonal variations of solar radiation is possible using Input Dynamics.
The effect of shading in the water column due to influent particulate material, as well as algae itself, is accounted for using the parameter "TSS concentration for calculating light extinction switching factor". When the simulated TSS concentration in the water column approaches this value, the growth rate of algae switches to zero regardless of what the specified value is for "Solar radiation for algal photosynthesis - depth averaged".
Growth of algae is accompanied by assimilation of ammonia as a nutrient source unless ammonia becomes limiting in which case nitrate is assimilated. Phosphorus and carbon dioxide are also assimilated for growth.
Respiration of algae is simulated when oxygen is available. Respiration means the conversion of algae into water, CO2, ammonia and phosphate by oxydation using oxygen. In the absence of oxygen, the respiration rate switches to zero and decay of algae is switched on. Decay means the lysis of algae into particulate organic material, ammonia and phosphate and occurs in the pond sediments where oxygen is absent.
¶ Mechanisms of focus models
The chapter describes in details the model mechanisms and refers to publications.
¶ Sumo2C
The whole-plant model SUMO2 (Dynamita, 2016, described in previous chapter) considering typical biological and physio-chemical model was modified to include the required components and processes in accordance with the critical review of existing models and experimental data, as summarized in Sumo2C Figure 1:
- Readily biodegradable substrate (SB) is split into monomers and polymers (SB,mono and SB,poly).
- A new biomass, A-Stage Heterotrophic Organisms (AHO) is added. This biomass stores SB,mono and SVFA into XSTO and has a high growth rate, while the Ordinary Heterotrophic Organisms consume only SB,poly and have a reduced growth rate.
- Hydrolysis process is performed only by low growth rate heterotrophic organisms (OHO, MEOLO, PAO and GAO) to simulate the low hydrolysis observed in A-Stage processes.
- Decay processes produce a fraction (fC) of colloids, split into biodegradable and unbiodegradable colloids (CB and CU) in the same fE proportion, as XE (particulate endogenous decay products) and XB,e (slowly biodegradable substrate) are generated in death-regeneration models.
- The modified model incorporates modeled EPS (XMEPS) as a flocculation agent, calculated variable based on a weighted sum of biomasses (XBIO) and particulate endogenous decay products (XE) related to the particulate COD (XCOD), with a saturation function on oxygen, meaning that under low DO, EPS are less produced (Nogaj et al., 2015). The calculated XMEPS is an indicator of EPS production, but the calculated values results depending on the analysis method used by different authors.
- The kinetic rate of flocculation processes are first order reactions that depend on the calculated XMEPS concentration. The rate is reduced under anoxic and anaerobic conditions to match with experimental observations. Furthermore, the flocculation rate is sensitive to temperature and to mixing intensity, represented in the model with a flocculation factor (Ff in %) that can be vary in each reactor depending on the aeration and mixing technology (Ff=0.1 for mechanical mixing, 0.5 for coarse bubbles, 0.75 for anaerobic processes).
- An empirical pump process unit is implemented to simulate the deflocculation processes occurring at pumping. The factor for the increase in CB/XB and CU/XU ratio can be adjusted depending on the pump technology.
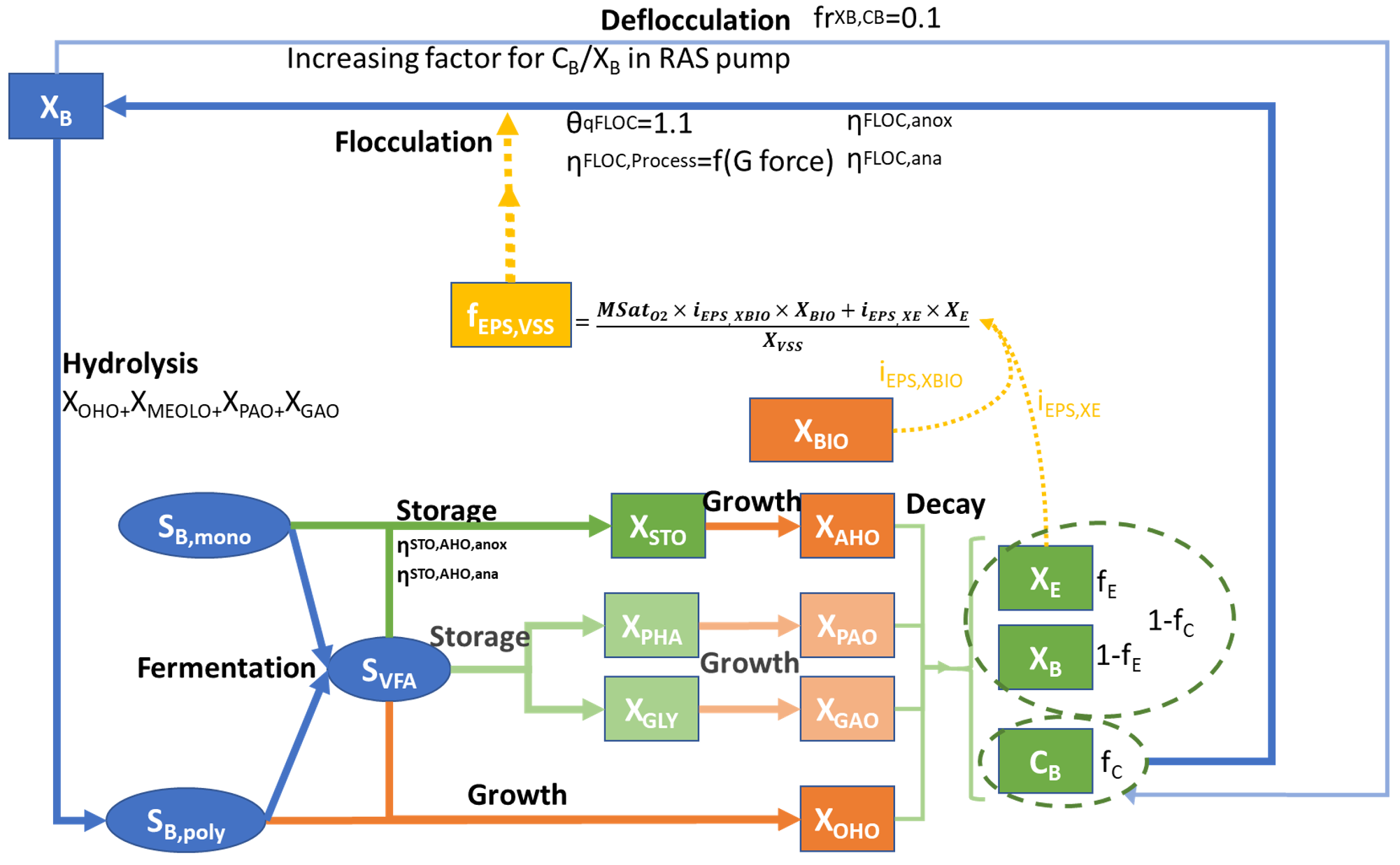
¶ Carbon absorption heterotroph organisms, XAHO
Heterotrophic organisms storing readily biodegradable small molecular weight components represented as SB,mono and volatile fatty acids (SVFA) into XSTO. The growth rate of AHOs is higher than that of the OHOs and outgrow them in a short SRT system of <2 days. They only carry out aerobic consumption of the XSTO and are expected to be seeded from the influent.
| Biological processes | Concepts description |
|---|---|
| AHO storage of SB,mono | Storage of SB,mono into XSTO without any energy required |
| AHO storage of SVFA | Storage of SVFA into XSTO without any energy required |
| AHO growth on XSTO, O2 | Requires O2, XSTO and nutrients. This is the AHO growth process. |
| AHO decay | Decay process of AHO under anoxic and aerobic conditions, resulting in XB,e, CB and XE release. XSTO is also considered to be released in the proportion of XSTO/XAHO. |
| AHO anaerobic decay | Decay process of AHO under anaerobic conditions, resulting in XB,e, CB and XE,ana release. XSTO is also considered to be released in the proportion of XSTO/XAHO. |
Haider, S., Svardal, K., Vanrolleghem, P.A., Kroiss, H., 2003. The effect of low sludge age on wastewater fractionation (S(S), S(I)). Water Sci. Technol. J. Int. Assoc. Water Pollut. Res. 47, 203–209.
Nogaj, T., Randall, A., Jimenez, J., Takacs, I., Bott, C., Miller, M., Murthy, S., Wett, B., 2015. Modeling of organic substrate transformation in the high-rate activated sludge process. Water Sci. Technol. 71, 971–979. https://doi.org/10.2166/wst.2015.051
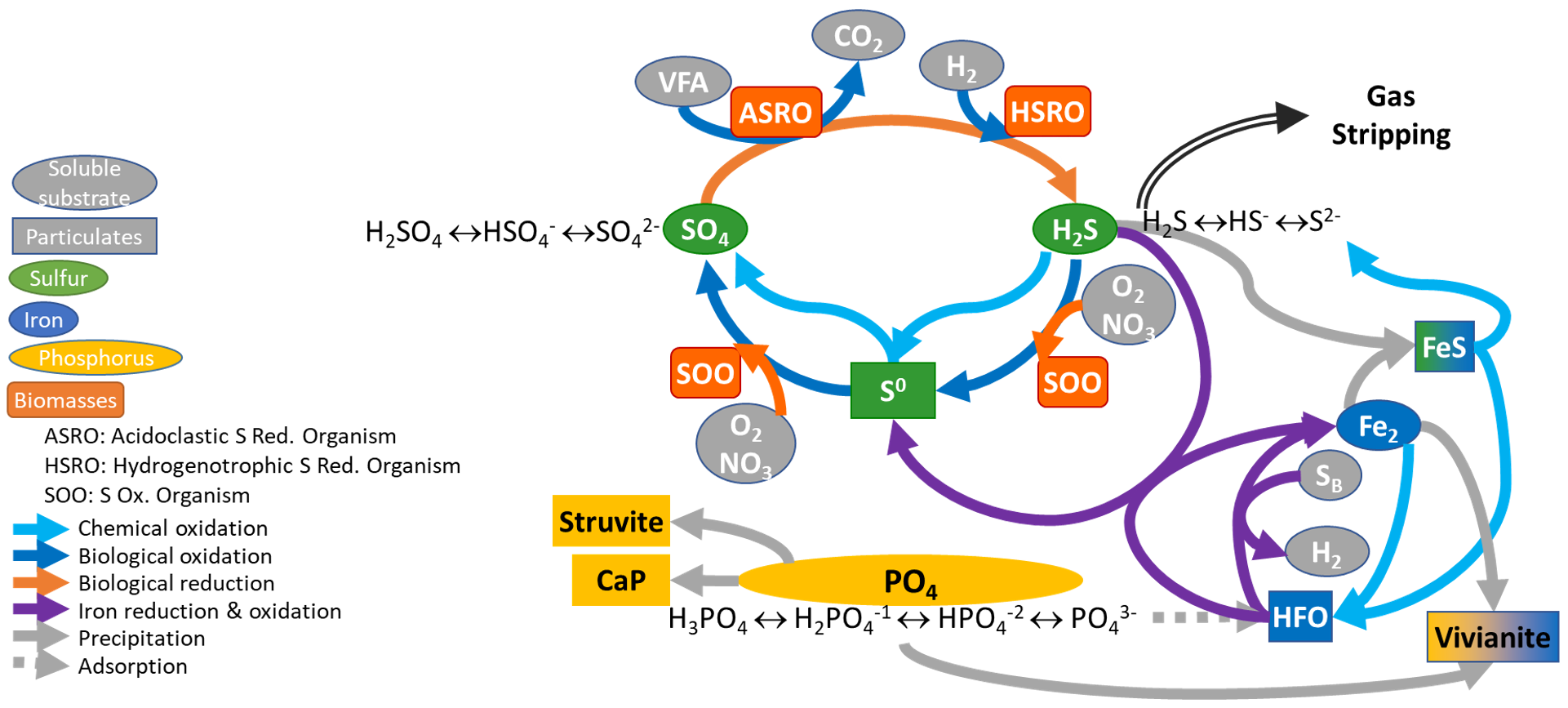
¶ Sumo2S
Biology and chemistry was extended with required components, species and reactions in accordance to the following literature review. Sumo2S Figure 1 synthesizes the sulfur cycle implemented in Sumo© models (Dynamita 2018) and the interactions with phosphorus and iron cycles.
¶ General sulfur and iron model
The sulfur model included in Sumo includes three oxidation states of sulfur: Sulfate (SO4-2) as SSO4, elemental sulfur (S0) as XS, and sulfide (S-) as SH2S.
Considering iron, two oxidation states are included in the model. Hydrous ferric oxides (HFO) species are already included in the base model for the chemical phosphorus treatment. These state variables (XHFO,H, XHFO,L, XHFO,old, XHFO,H,P, XHFO,L,P, XHFO,H,P,old and XHFO,L,P,old depending on the floc size and P-bound status) are considered to be the only ferric (Fe3+) species in the model, as ferric iron is only minimally soluble in water (Hauduc et al., 2015). XHFO is a calculated variable being the sum of the seven HFO state variables. For the ferrous iron (Fe2+), a new state variable is included in the model as SFe2, and ferrous oxides are not considered.
These states are considered to interact with other wastewater components as described below.
¶ FeS precipitation and iron interaction
¶ Reduction of Fe3+, sulfide as electron donor
A chemical reduction of Fe3+ by sulfide occurs under reducing conditions. In this reaction, sulfide is oxidized into colloidal elemental sulfur which precipitates (Firer et al., 2008; Nielsen et al., 2005):
2 Fe3+ + HS- → 2 Fe2+ + S0 + H+
Implementation in the whole plant model: Elemental sulfur has been added as a particulate state variable (XS), as elemental sulfur has a low solubility and flocculates easily. The hydrous ferric oxides (XHFO) are reduced by H2S in a single process into ferrous iron (SFe2) and elemental sulfur (XS) with adequate stoichiometric coefficient to balance the redox reaction and a first order rate with respect to the XHFO concentration.
¶ Reduction of Fe3+, organic matter as electron donor
Hydrous ferric oxides are reduced in digesters into soluble Fe2+ which precipitates into iron sulfide [FeS], and release bounded phosphates (Ge et al., 2013), that can further precipitate into vivianite [Fe3(PO4)2,8H2O] (Cheng et al., 2015). This biological process is performed by Fe3+ reducing bacteria, using organic matter as electron donors (Lovley and Phillips, 1988).
Implementation in the whole plant model: To keep the model simple, this new iron reducing biomass and the associated growth is not introduced in the model. The soluble biodegradable substrate (SB) and volatile acids (SVFA) are considered as electron donor and a first order kinetic rate expression with respect to the hydrous ferric oxide (XHFO) concentration is used.
¶ FeS precipitation
Fe2+ precipitates with sulfide into iron sulfide, FeS (Firer et al., 2008; Nielsen et al., 2005):
Fe2+ + HS- → FeS + H+
Implementation in the whole plant model: The acid-base reactions of the sulfate and sulfide species are added in the pH model for speciation (equilibrium model). The precipitation is modelled following Koutsoukos et al (1980) kinetic expression with a solubility product Ksp,FeS=3.7*10-19 (Nielsen et al., 2005).
¶ Oxidation of Fe2+
According to Gutierrez et al. (2010), the precipitated iron sulfide (FeS) is re-oxidized into ferric oxides and sulfate in an aerobic zone.
Implementation in the whole plant model: Both oxidation of ferrous iron (SFe2) and precipitated iron sulfide (XFeS) are considered in the model with oxygen as electron acceptor with adequate stoichiometric coefficient to balance the redox reaction and a first order rate with respect to the SFe2 and XFeS concentrations respectively.
¶ Reduction of sulfate
The biological sulfate reduction is the main process step in sulfur biotreatment, often combined with a chemical step or a metal precipitation step (Hao et al., 2014). The biological sulfate reduction is performed by sulfate reducing organisms (SRO), which can use either hydrogen or organic compounds as electron donor. These bacteria are directly in competition with hydrogenotrophic and acetoclastic methanogens respectively in anaerobic bioprocesses (Chou et al., 2008; Hao et al., 2014; Kalyuzhnyi and Fedorovich, 1998) and in sewer sediments (Liu et al., 2016). Models for sewer system usually neglect the biomass growth whereas models for anaerobic digestion always consider it. These models consider different kind of substrates. Knobel and Lewis (2002), Liu et al. (2015) and Fedorovich et al. (2003) consider 5 or 4 substrates respectively (different volatile fatty acids and H2), whereas Batstone (2006) suggests considering only hydrogenotrophic sulfate reducer bacteria if S/COD ratio is below 0.1 mg S/mg COD. The best compromise seems to be the model from Barrera et al. (2015) and Poinapen and Ekama (2010) who consider H2, acetate and propionate as substrates. The WATS model for sewer processes (Hvitved-Jacobsen et al., 2013) consider only soluble substrate for sulfate reduction biological processes.
Implementation in the whole plant model: Considering the actual structure of the extended version of Sumo model, SVFA and SH2 have been chosen as substrate for sulfate reducing organisms (SRO), resulting in competition with the AMETO and HMETO, which would be similar to what is suggested by Barrera (2015) and in accordance with Kalyuzhnyi and Federovich (1998). Similarly to the methanogenesis implementation, two biomasses are introduced: ASRO (Acidoclastic Sulfate-Reducing Organisms) and HSRO (Hydrogenotrophic Sulfate-Reducing Organisms). This leads to 4 additional processes to consider growth and decay of both biomasses. Stoichiometric and kinetic values from Barrera (2015) are used. The produced sulfide is inhibitory (Utgikar et al. 2002). It has been considered in the kinetic rate expression through Haldane functions when sulfide is a reactant of the process, otherwise through Monod limitation function term.
¶ Oxidation of sulfide
¶ Biological oxidation
The biological oxidation of sulfide into sulfate is performed through intermediate species. The oxidation may use either oxygen, nitrite or nitrate as electron acceptor. In the literature, the biological oxidation of sulfide is mainly modelled in one or two steps, elemental sulfur (S0) being the intermediate. The oxidation of elemental sulfur to sulfate is the limiting step (Buisman, et al., 1991; Jiang et al., 2009; Tichy et al., 1998). According to several authors, when sulfide is oxidized in a digester at limited oxygen levels, it reacts to elemental sulfur which precipitates, making it less available for further biological reduction (Diaz and Fdz-Polanco 2012; Jenicek et al. 2008).
Implementation in the whole plant model: A Sulfur Oxidizing Organism (XSOO) has been introduced in the model with four oxidation processes to consider the two steps of sulfide oxidation and two possible oxidants (O2 and NO3). The parameter values from Mannucci et al (2012) are used as first estimation.
¶ Chemical oxidation
At high SOO activity, chemical oxidation is negligible (Luther et al., 2011) but must be considered in case of sewer processes with lower biomass concentration as the oxygen consumption for sulfide oxidation count significantly in the OUR (Nielsen et al., 2003). The literature reports kinetic laws with different orders and a wide range of oxidation rate parameters, however the rate of the two steps of oxidation are not determined independently (Buisman et al., 1990; Hvitved-Jacobsen et al., 2013; Klok et al., 2013; Luther et al., 2011; Nielsen et al., 2003).
Implementation in the whole plant model: Two processes for oxidation of SH2S by oxygen in two steps (SH2S→XS→SSO4) is added. All the oxidation intermediates are considered through the elemental sulfur state variable (XS), whereas the second oxidation step (XS→SSO4) is much slower (Nielsen et al., 2003). To simplify the model, first order reactions with respect to sulfide and to elemental sulfur has been implemented for both steps of the oxidation process.
¶ Biomasses involved in sulfur reactions
¶ Acidoclastic sulfate-reducing organisms, XASRO
A group of bacteria and archaea that perform anaerobic respiration utilizing SSO4 and SVFA, reducing it to SH2S and generating SCO2. They compete with AMETOs for SVFA and negatively impact performance of a digester.
| Biological processes | Concepts description |
|---|---|
| ASRO growth - SO4 reduction with SVFA |
ASRO growth process on SVFA, using SSO4 as electron acceptor. SH2S is produced. Requires SSO4 and nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH. |
| ASRO decay | ASRO decay process under anoxic and aerobic conditions. This process release XB,e and XE (death-regeneration concept). |
| ASRO anaerobic decay | ASRO decay process under anaerobic conditions. This process release XB,e and XE,ana (death-regeneration concept). |
¶ Hydrogenotrophic sulfate-reducing organisms, XHSRO
A group of bacteria and archaea that perform anaerobic respiration utilizing SSO4 and SH2, reducing it to SH2S. They compete with HMETOs for SH2 consumption and negatively impact performance of a digester.
| Biological processes | Concepts description |
|---|---|
| HSRO growth - SO4 reduction with SH2 |
HSRO growth process on SH2, using SSO4 as electron acceptor. SH2S is produced. Requires SSO4 and nutrients (SNHx, SPO4, SCAT, SAN, SCa, SMg). Bell shape inhibition function on pH: BellinhpH. |
| HSRO decay | HSRO decay process under anoxic and aerobic conditions. This process release XB,e and XE (death-regeneration concept). |
| HSRO anaerobic decay | HSRO decay process under anaerobic conditions. This process release XB,e and XE,ana (death-regeneration concept). |
¶ Sulfur-oxidizing organisms, XSOO
Sulfur-oxidizing organisms (SOO) oxidize SH2S in two steps under aerobic and anoxic environments, first from SH2S to XS (elemental sulfur) and second from XS to SSO4.
| Biological processes | Concepts description |
|---|---|
| SOO growth on H2S, O2 |
SOO aerobic growth, using SH2S as electron donor. SH2S is oxidised into elemental sulfur XS with SO2 as electron acceptor. Requires SH2S, SO2 and nutrients (SH2S, SNHx, SPO4, SCAT, SAN, SCa, SMg). |
| SOO growth on H2S, NO2 |
SOO anoxic growth, using SH2S as electron donor. SH2S is oxidised into elemental sulfur XS with SNO2 as electron acceptor. Requires SH2S, SNO2 and nutrients (SH2S, SNHx, SPO4, SCAT, SAN, SCa, SMg). |
| SOO growth on H2S, NO3 |
SOO anoxic growth, using SH2S as electron donor. SH2S is oxidised into elemental sulfur XS with SNO2 as electron acceptor. Requires SH2S, SNO2 and nutrients (SH2S, SNHx, SPO4, SCAT, SAN, SCa, SMg). |
| SOO growth on S°, O2 |
SOO aerobic growth, using elemental sulfur XS as electron donor. XS is oxidised into sulfate SSO4 with SO2 as electron acceptor. Requires XS, SO2 and nutrients (SH2S, SNHx, SPO4, SCAT, SAN, SCa, SMg). |
| SOO growth on S°, NO2 |
SOO anoxic growth, using elemental sulfur XS as electron donor. XS is oxidised into sulfate SSO4 with SNO2 as electron acceptor. Requires XS, SNO2 and nutrients (SH2S, SNHx, SPO4, SCAT, SAN, SCa, SMg). |
| SOO growth on S°, NO3 |
SOO anoxic growth, using elemental sulfur XS as electron donor. XS is oxidised into sulfate SSO4 with SNO3 as electron acceptor. Requires XS, SNO3 and nutrients (SH2S, SNHx, SPO4, SCAT, SAN, SCa, SMg). |
| SOO decay | SOO decay process under anoxic and aerobic conditions. This process release XB,e and XE (death-regeneration concept). |
| SOO anaerobic decay | SOO decay process under anaerobic conditions. This process release XB,e and XE,ana (death-regeneration concept). |
Barrera, E. L., Spanjers, H., Solon, K., Amerlinck, Y., Nopens, I., and Dewulf, J. (2015) Modeling the anaerobic digestion of cane-molasses vinasse: Extension of the Anaerobic Digestion Model No. 1 (ADM1) with sulfate reduction for a very high strength and sulfate rich wastewater. Water Research, 71, 42–54.
Batstone, D. J. (2006) Mathematical Modelling of Anaerobic Reactors Treating Domestic Wastewater: Rational Criteria for Model Use. Reviews in Environmental Science and Bio/Technology, 5(1), 57–71.
Buisman, C., Ijspeert, P., Hof, A., Janssen, A., Tenhagen, R., and Lettinga, G. (1991) Kinetic-Parameters of a Mixed Culture Oxidizing Sulfide and Sulfur with Oxygen. Biotechnology and Bioengineering, 38(8), 813–820.
Buisman, C., Uspeert, P., Janssen, A., and Lettinga, G. (1990) Kinetics of chemical and biological sulphide oxidation in aqueous solutions. Water Research, 24(5), 667–671.
Chou, H.-H., Huang, J.-S., Chen, W.-G., and Ohara, R. (2008) Competitive reaction kinetics of sulfate-reducing bacteria and methanogenic bacteria in anaerobic filters. Bioresource Technology, 99(17), 8061–8067.
Díaz, I. & Fdz-Polanco, M. 2012 Robustness of the microaerobic removal of hydrogen sulfide from biogas. Water Science and Technology 65, 1368–1374.
Fedorovich, V., Lens, P., and Kalyuzhnyi, S. (2003) Extension of Anaerobic Digestion Model No. 1 with Processes of Sulfate Reduction. Applied Biochemistry and Biotechnology, 109(1-3), 33–46.
Firer, D., Friedler, E., and Lahav, O. (2008) Control of sulfide in sewer systems by dosage of iron salts: Comparison between theoretical and experimental results, and practical implications. Science of the Total Environment, 392(1), 145–156.
Gutierrez, O., Park, D., Sharma, K. R., and Yuan, Z. (2010) Iron salts dosage for sulfide control in sewers induces chemical phosphorus removal during wastewater treatment. Water Research, 44(11), 3467–3475.
Hao, T., Xiang, P., Mackey, H. R., Chi, K., Lu, H., Chui, H., van Loosdrecht, M. C. M., and Chen, G.-H. (2014) A review of biological sulfate conversions in wastewater treatment. Water Research, 65, 1–21.
Hauduc, H., Takács, I., Smith, S., Szabo, A., Murthy, S., Daigger, G.T., Spérandio, M., 2015. A dynamic physico-chemical model for chemical phosphorus removal. Water Research. 73, 157–170.
Hvitved-Jacobsen, T., Vollertsen, J., and Nielsen, A. H. (2013) Sewer Processes: Microbial and Chemical Process Engineering of Sewer Networks, Second Edition, CRC Press.
Jenicek, P., Keclik, F., Maca, J. & Bindzar, J. 2008 Use of microaerobic conditions for the improvement of anaerobic digestion of solid wastes. Water Science and Technology 58,1491–1496.
Jiang, G., Sharma, K. R., Guisasola, A., Keller, J., and Yuan, Z. (2009) Sulfur transformation in rising main sewers receiving nitrate dosage. Water Research, 43(17), 4430–4440.
Kalyuzhnyi, S. V. and Fedorovich, V. V. (1998) Mathematical modelling of competition between sulphate reduction and methanogenesis in anaerobic reactors. Bioresource Technology, 65(3), 227–242.
Klok, J. B. M., de Graaff, M., van den Bosch, P. L. F., Boelee, N. C., Keesman, K. J., and Janssen, A. J. H. (2013) A physiologically based kinetic model for bacterial sulfide oxidation. Water Research, 47(2), 483–492.
Knobel, A. N. and Lewis, A. E. (2002) A mathematical model of a high sulphate wastewater anaerobic treatment system. Water Research, 36(1), 257–265.
Liu, Y., Tugtas, A. E., Sharma, K. R., Ni, B.-J., and Yuan, Z. (2016) Sulfide and methane production in sewer sediments: Field survey and model evaluation. Water Research, 89, 142–150.
Liu, Y., Zhang, Y., and Ni, B.-J. (2015) Evaluating Enhanced Sulfate Reduction and Optimized Volatile Fatty Acids (VFA) Composition in Anaerobic Reactor by Fe (III) Addition. Environmental Science & Technology, 49(4), 2123–2131.
Luther, G. W., Findlay, A. J., MacDonald, D. J., Owings, S. M., Hanson, T. E., Beinart, R. A., and Girguis, P. R. (2011) Thermodynamics and Kinetics of Sulfide Oxidation by Oxygen: A Look at Inorganically Controlled Reactions and Biologically Mediated Processes in the Environment. Frontiers in Microbiology, 2:62.
Mannucci, A., Munz, G., Mori, G., Lubello, C., 2012. Biomass accumulation modelling in a highly loaded biotrickling filter for hydrogen sulphide removal. Chemosphere 88, 712–717.
Nielsen, A. H., Lens, P., Vollertsen, J., and Hvitved-Jacobsen, T. (2005) Sulfide-iron interactions in domestic wastewater from a gravity sewer. Water Research, 39(12), 2747–2755.
Nielsen, A. H., Vollertsen, J., and Hvitved-Jacobsen, T. (2003) Determination of kinetics and stoichiometry of chemical sulfide oxidation in wastewater of sewer networks. Environmental Science & Technology, 37(17), 3853–3858.
Poinapen, J. and Ekama, G. A. (2010) Biological sulphate reduction with primary sewage sludge in an upflow anaerobic sludge bed reactor - Part 5: Steady-state model. Water SA, 36(3), 193–202.
Tichy, R., Janssen, A., Grotenhuis, J. T. C., Van Abswoude, R., and Lettinga, G. (1998) Oxidation of biologically-produced sulphur in a continuous mixed-suspension reactor. Water Research, 32(3), 701–710.
¶ Sumo4N
To identify N2O formation pathways on various wastewater treatment processes, several attempts to describe nitrification and denitrification intermediates in dynamic models have been published in the last decade. However, there is no clear consensus on the N2O production modelling concepts yet. The combination of the 4-step denitrification model from Hiatt & Grady (2008) and the 2-P model for ammonium oxidizing bacteria from Pocquet et al. (2016) has been used in recent studies and seems promising (Mampaey et al, 2019; Fiat et al., 2019).
In Sumo24, abiotic reactions of hydroxylamine oxidation to N2O have been introduced to solve a model conceptual problem, as described below.
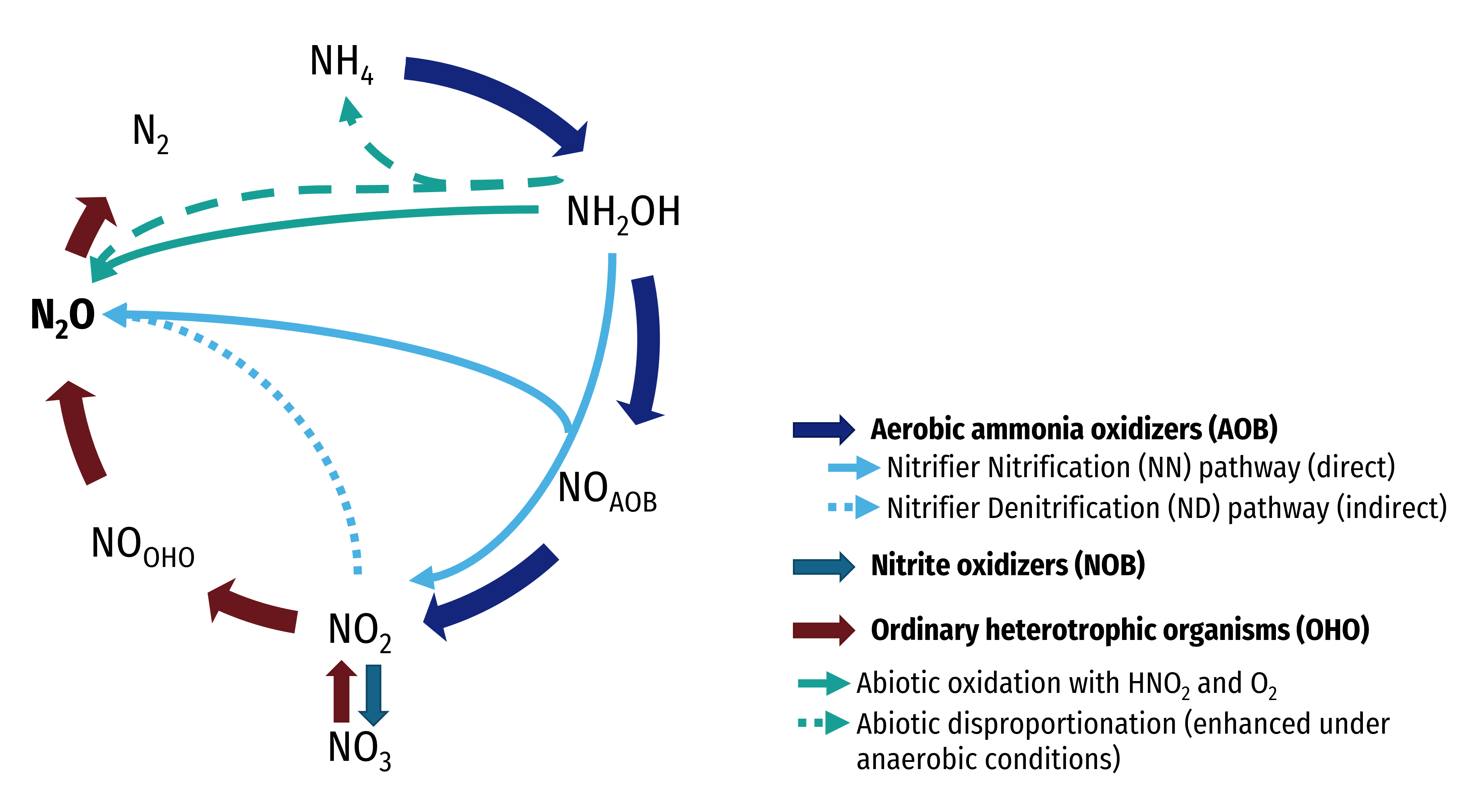
What we know about aerobic N2O production so far:
- The contribution of ND pathway is usually higher than the contribution of NN pathway
- The contribution of ND pathway increases at low DO under 1 mg O2/L (Peng et al.,2014).
- The contribution of the ND pathway increases with high nitrite at low DO level (Wunderlin et al., 2013).
- The contribution of NN pathway increases with the DO (Peng et al.,2014).
Hydroxylamine inhibition on NOB is added in accordance with Jofra-Soler et al (2021).
¶ Abiotic reactions of hydroxylamine
Hydroxylamine is an intermediate compound of nitrification, but not of heterotrophic denitrification metabolism (Soler-Jofra et al., 2021). In the model from Pocquet et al. (2016), the hydroxylamine can only be oxidized by AOBs with oxygen or nitrite. The application of this model in a wide range of layouts shows that hydroxylamine could be accumulated to a few g/m3 and remains unchanged in anaerobic reactors when there is internal recirculation. The presence of hydroxylamine under anaerobic conditions was causing some discrepancies in the ORP calculation compared to other Sumo models, and had thus an impact on BioP efficiency (see Interactions with BioP model)
Although a lot of unknown remains around hydroxylamine metabolism, it is an inorganic, highly reactive compound that is intermediate or side metabolite in different nitrogen cycle microorganisms (Soler-Jofra et al., 2021). Consequently, it is unlikely that it is unchanged under anaerobic conditions.
To cope with this issue and provide more flexibility in the calibration of N2O production under a wide range of conditions, it was chosen to implement the 3 main abiotic reactions that involve hydroxylamine (Soler-Jofra et al., 2021; Su et al., 2019; Ye et al., 2022):
- Abiotic oxidation of hydroxylamine with nitrite with a rate equation as a function of pH, SNO2 and SNH2O2 (Su et al., 2019)
- Abiotic oxidation of hydroxylamine with oxygen, with a low rate parameter as no reliable value has been published in the literature to our knowledge
- Abiotic disproportionation into NHx and N2O is added as described in Su et al. (2019) but an increased rate under anaerobic conditions is added to deplete hydroxylamine under anaerobic conditions. This can be seen as a lumped process for all the metabolism pathways for hydroxylamine under anaerobic conditions that are not yet well described in the literature.
The other abiotic reactions are not implemented yet as their contribution seems to have less importance.
¶ 4-step denitrification: Anoxic growth of OHOs from Hiatt&Grady (2008, only for OHO denitrification on SB and SVFA)
- Reduction factor for anoxic growth from Hiatt&Grady, 2008
- Half-saturation for substrates and O2 kept at Sumo2 values
- Half saturation for NO3 and NO2 from Hiatt&Grady2008
- Same anoxic yield than in Hiatt&Grady (0.54), although the aerobic yield in Sumo is higher than in Hiatt&Grady (0.67 and 0.54 respectively)
- In Sumo2 inhibition terms on NO2 make sure that the denitrification reactions are occurring successively. These terms are not used in Hiatt&Grady and have been removed.
- Assimilative nitrate/nitrite reduction to ammonia coded differently in Sumo2 (and kept as is): NO3-→NH4+ and NO2-→NH4+ directly, biomasses are used as electron donors instead of substrate
- Mixotrophic growth of NOB not implemented
These parameters take into consideration the fraction of heterotrophic bacteria accomplishing each step of denitrification and the reduced maximum specific growth rate under anoxic conditions.
This results in 8 denitrification processes:
| 2 | r2 | OHO growth on VFAs, NO3 |
| 3 | r3 | OHO growth on VFAs, NO2 |
| 4 | r4 | OHO growth on VFAs, NO |
| 5 | r5 | OHO growth on VFAs, N2O |
| 7 | r7 | OHO growth on SB, NO3 |
| 8 | r8 | OHO growth on SB, NO2 |
| 9 | r9 | OHO growth on SB, NO |
| 10 | r10 | OHO growth on SB, N2O |
Growth processes on volatile fatty acids (SVFA) or readily biodegradable substrate (SB) are similar and occur successively thanks to the switch functions:
- MsatVFA,KVFA used for growth on VFA processes
- MsatSB,KSB*MinhSVFA,KVFA used for growth on readily biodegradable processes
¶
| Biological processes | Concepts description |
|---|---|
|
OHO growth with NO3
|
Reduction NO3-→NO2- Kinetic parameters
Kinetic rate limitation/inhibitions
|
|
OHO growth with NO2
|
Reduction NO2-→NO,OHO (NO considered as cell-internal intermediate) Kinetic parameters
Kinetic rate limitation/inhibitions
|
|
OHO growth with NO
|
Reduction NO,OHO→N2O (NO considered as cell-internal intermediate) Kinetic parameters
Kinetic rate limitation/inhibitions
|
|
OHO growth with N2O
|
Reduction N2O→N2 Kinetic parameters
Kinetic rate limitation/inhibitions
|
¶ 4-step AOB nitrification: From Pocquet et al. 2016
Five processes are included in the model. Nitric oxide (NO) is considered to be a metabolism intermediate and thus unlikely being released in the bulk and stripped. Consequently, the correction factor for mass transfer of NO parameter (fkL,GNO) is set to zero.
- AOB decay kept as in Sumo2
- NOB growth and decay kept as in Sumo2
| Biological processes | Concepts description |
|---|---|
|
AOB NHx oxidation to NH2OH
|
Requires O2. The Monod term in the original model (Pocquet et al, 2016) is on free ammonia. It has been replaced by a term on total ammonia for more stability of the model under normal pH range. Kinetic parameters The maximum rate is the maximum specific growth rate of AOBs divided by the AOB yield (µAOB/YAOB) to keep the same substrate utilization rate than for the AOB growth process (AOB NH2OH oxidation to NO process)
Kinetic rate limitation/inhibitions
|
|
AOB NH2OH oxidation to NO
|
Requires O2. This is the AOB growth process. Kinetic parameters
Kinetic rate limitation/inhibitions
|
|
AOB NO oxidation to NO2
|
Requires O2. Kinetic parameters The maximum rate is the maximum specific growth rate of AOBs divided by the AOB yield (µAOB/YAOB) to keep the same substrate utilization rate than for the AOB growth process (AOB NH2OH oxidation to NO process)
KNO,NO2,AOB,AS: Half-saturation of NO to NO2 for AOBs (AS) Kinetic rate limitation/inhibitions
|
|
AOB NO reduction to N2O (NN pathway)
|
Direct pathway of N2O production (or NN pathway): NO reduction to N2O by the enzyme “Nor” coupled with the hydroxylamine oxidation to nitrite Kinetic parameters The maximum rate is the maximum specific growth rate of AOBs divided by the AOB yield (µAOB/YAOB), reduced by a reduction factor for NO reduction to N2O by AOBs (NN pathway) (ƞAOB,NO,N2O)
Kinetic rate limitation/inhibitions
|
| AOB HNO2 reduction to N2O (ND pathway) |
Indirect pathway of N2O production (or ND pathway): HNO2 reduction to N2O coupled with NH2OH oxidation to nitrite. Originally this ND pathway considers 2 steps: NO2 reduction to NO (NirK enzyme), then NO reduction to N2O (Nor enzyme) (Mampaey et al., 2013; Ni et al., 2011). These two processes are merged in this single one to avoid the NO loop with the AOB NO oxidation to NO2 process (Pocquet et al., 2016). the Haldane-type term on O2 is replaced with a simple Monod inhibition term. This results in a limitation of this process under anoxic conditions with hydroxylamine, what is observed by some authors (Domingo-Félez and Smets, 2016) Stoichiometric parameters
Kinetic parameters The maximum rate is the maximum specific growth rate of AOBs divided by the AOB yield (µAOB/YAOB), reduced by a reduction factor for HNO2 reduction to N2O by AOBs (ND pathway) (ƞAOB,NO2,N2O)
Kinetic rate limitation/inhibitions
|
¶ Interactions with BioP model
Sumo4N model is expected to behave as Sumo2 model for overall nitrification and denitrification processes, and for all other biological processes including biological phosphorus removal. The BioP efficiency is highly dependent on ORP calculation (link to ORP chapter), which drives the PAO/GAO ratio (see BioP model description). Consequently, it is important to reproduce similar ORP calculations with Sumo4N as in Sumo2. In Sumo2, anoxic ORP calculation is based on SNOx, which is the sum of nitrate and nitrite. To get similar values of ORP under anoxic conditions with Sumo4N, it is required to account for other nitrification intermediates (SNOx,Tot=SNO2 + SNO3 + SNO + SNH2OH).
This modification in the anoxic ORP calculation explains the importance of depleting hydroxylamine under anaerobic conditions to allow similar ORP values compared to Sumo2.
Notes about the steady-state solver
The “Fast” solver may fail to find the proper solution under certain circumstances. In this case, we advise to first run few days dynamically, or use the “accurate” solver.
Fiat, J., Filali, A., Fayolle, Y., Bernier, J., Rocher, V., Spérandio, M., Gillot, S., 2019. Considering the plug-flow behavior of the gas phase in nitrifying BAF models significantly improves the prediction of N2O emissions. Water Res. 156, 337–346. https://doi.org/10.1016/j.watres.2019.03.047
Hiatt, W.C., Grady, C.P.L. Jr., 2008. An updated process model for carbon oxidation, nitrification, and denitrification. Water Environ. Res. 80(11), 2145-2156.
Houweling, Dwight; Constantine, Tim; Eriksen Søren; Uri Carreño, Nerea; Holmen Andersen, Mikkel. 2016. N2O Emissions from Sidestream Deammonification: Using Latest Generation Models to Guide Optimization. 2016 WEFTEC Proceedings
Ni, B.-J., Law, Y., Guo, J., Yuan, Z. 2014. Modeling of Nitrous Oxide Production by Autotrophic Ammonia-Oxidizing Bacteria with Multiple Production Pathways. Environmental Science and Technology, 48(7):3916-24.
Mampaey, K.E., Spérandio, M., van Loosdrecht, M.C.M., Volcke, E.I.P., 2019. Dynamic simulation of N2O emissions from a full-scale partial nitritation reactor. Biochem. Eng. J. 152, 107356. https://doi.org/10.1016/j.bej.2019.107356
Pocquet, M., Wu, Z., Queinnec, I., Spérandio, M., 2016. A two pathway model for N2O emissions by ammonium oxidizing bacteria supported by the NO/N2O variation. Water Res. 88, 948-959.
Soler-Jofra, A., Pérez, J., van Loosdrecht, M.C.M., 2021. Hydroxylamine and the nitrogen cycle: A review. Water Res. 190, 116723. https://doi.org/10.1016/j.watres.2020.116723
Su, Q., Domingo-Félez, C., Jensen, M.M., Smets, B.F., 2019. Abiotic Nitrous Oxide (N2O) Production Is Strongly pH Dependent, but Contributes Little to Overall N2O Emissions in Biological Nitrogen Removal Systems. Environ. Sci. Technol. 53, 3508–3516. https://doi.org/10.1021/acs.est.8b06193
Uri, Nerea; Nielson, Per H.; Holmen Andersen, Mikkel; Hafner, Sasha; Li, Zheqin; Chandran, Kartik. 2017. Continuous Aeration Control to Reduce N2O Emissions in a Full-Scale Sidestream Deammonification Reactor. 2017 WEFTEC Proceedings
Ye, L., Porro, J., Nopens, I. (Eds.), 2022. Quantification and Modelling of Fugitive Greenhouse Gas Emissions from Urban Water Systems. IWA Publishing. https://doi.org/10.2166/9781789060461
¶ Physico-chemical processes
¶ pH and alkalinity
The first mechanistic models (e.g. ASM1) did alkalinity accounting to estimate running out of alkalinity and the detrimental effect on nitrification through lack of bicarbonate and low pH. Starting from ADM1 and all modern software today calculate pH instead based on chemical equilibrium and determine alkalinity by "virtual" titration.
There are three different methods to calculate pH:
1) Kinetic approach such as the Musvoto (2000) - this is easy to implement in existing Gujer matrix tools, but computationally very inefficient due to the large difference between the rates of protonation-deprotonation and biological reactions.
2) Tableau - this is a standard chemical method, easy to extend, but it requires solving a multidimensional non-linear problem involving key species. It is quite OK for determining pH of one solution, but when we have 1000 locations in the plant model, some within recycle loops etc., the problem dimension may easily become hundred thousand and unfeasible to solve efficiently every single timestep.
3) The one most software uses: Solve pH, alkalinity and ionic strength. If pH and ionic strength (IS) were known, each ionic species concentration can be calculated from the total component concentration (i.e. total ammonia+ammonium can be split at a certain pH and IS to undissociated ammonia and ammonium ions). The method is based on guessing a pH and IS value, calculating ionic species, checking the charge balance and the IS error, and using Newton-Raphson method to vary pH and IS until both charge balance and IS error are zero.
The advantage of this method is that the variables to be solved are limited (2 in the base case). Disadvantage is that the equations for species calculation must be developed manually for each species (though symbolic solvers are available such as Maple or SymPy Python library). After a certain complexity of the pH model (e.g. in the case of many species and ionic pairs) the equations can become unsolvable symbolically. In this case the problem dimension will be increased by adding one central species (such as Fe3+ or HCO3-) to pH and IS (so solving three variables instead of two). This drastically reduces the complexity of the equations and only marginally increases the solution time.
Consider a simple example for ammonia in distilled water, in a simple chemical model.
¶ Components definition
On the components tab of the model each state variable is defined and its total component concentrations (i.e. total ammonia + ammonium for SNHx) is setup in “Initial concentration” column. This is equivalent to the “Input setup” task of the GUI, see user manual for details.
¶ Speciation expressions
In a solution of pure ammonia, we have two dissociation/ionization reactions and a mass balance:
Water dissociation: [H+] * [OH-] = kW
Ammonia ionization: [H+] * [NH3] /[NH4+] = KiNH3
Mass balance: SNHx = [NH3] + [NH4+]
Three equations can be solved for three variables to calculate the speciation of ammonia:
[OH-] = kW /[H+]
[NH3] = (KiNH3 * SNHx) /([H+] + KiNH3)
[NH4+]= ([H+] * SNHx) /([H+] + KiNH3)
This can be solved through a symbolic solver, especially in case of more complex systems (carbonates, phosphates, ion pairing…). Sympy is a Python library for symbolic mathematics and can be used online (http://live.sympy.org/). The above system for ammonia speciation can be solved with the following code:
from sympy import symbols, solve, Eq
SNHx, OH, H, NH4_p, NH3, KiNH3, KW= symbols('SNHx, OH, H, NH4_p, NH3, KiNH3, KW')
solve((Eq(KW, H*OH), Eq( KiNH3, NH3*H/NH4_p), Eq(SNHx, NH4_p+NH3)), OH, NH4_p, NH3)
where
| from sympy import symbols, solve, Eq | Loads the appropriate libraries and functions |
| SNHx, OH, H, NH4_p, NH3, KiNH3, KW=symbols('SNHx, OH, H, NH4_p, NH3, KiNH3, KW') | Declares the variables and parameters |
| solve((Eq(KW, H*OH), Eq( KiNH3, NH3*H/NH4_p), Eq(SNHx, NH4_p+NH3)), OH, NH4_p, NH3) |
Provides the system of 3 equations (comma meaning “=” sign)
And the 3 unknown variables |
The results will be:
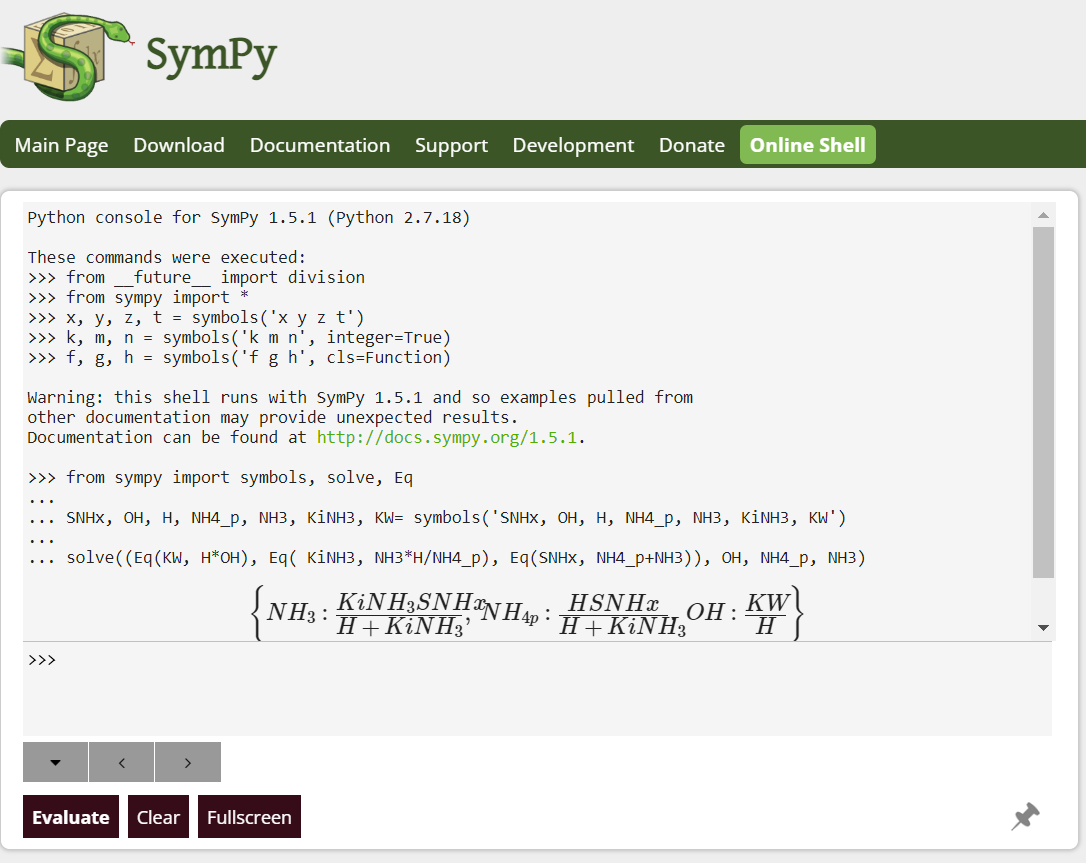
Knowing the definition of pH
[H+] = 10(-pH)
These equations are entered into the pH table in the Sumo model file, in the pH sheet:
SNHx needs to be converted from g/m3 (equivalent to mg/L, used in the kinetic model) to mol/L (used in the equilibrium model), therefore divided by the atomic mass of N (AMN * 1000 = 14000 mg/mol). Sumo constructs the necessary equation system and solver setup from this information automatically.
These species plotted as a function of pH provide the dissociation curves, we know (for 0.0001 molar pure ammonia solution):
It is possible to make a pH and ionic strength guess (i.e. pH=7.0, IS=0.0001 mol/kg)
¶ Dissociation and acidity constants
The dissociation and acidity constants used for the speciation calculations are sensitive to the temperature and ionic strength. They are corrected in the calculated variables of the model:
- For temperature
For water dissociation constants the Van’t Hoff equation is used to correct the constant value for temperature. For other compounds, empirical equations are used when available.
- For ionic strength
In a non-ideal aqueous phase, such as typical wastewater, the ions interact due to molecular attraction and repelling, and those interactions increase with the total charge concentration. These interactions reduce the number of ions that are available (active) for reactions. Therefore, the measured concentration must be corrected in order to get the activities the ions would have under ideal conditions.
Sumo is using Davies corrections equations for ionic strength, which is valid from ionic strength in the range of 0 – 0.5 mol/kg (Tait et al., 2012). The activity correction coefficient (fmono, fdi and ftri for monovalent, divalent and trivalent ions respectively) allows correcting ion concentrations for non-ideal aqueous phase.
In Sumo those activity coefficients are used to correct directly the dissociations constants, so that the calculated ion concentrations (in Species sheet) are directly the concentration measured by the operator, and this simplify the expressions in the equilibrium table.
As example, for water dissociation:
[H+]*fmono * [OH-]*fmono = KW
Is equivalent to:
[H+] * [OH-] = KW/fmono²
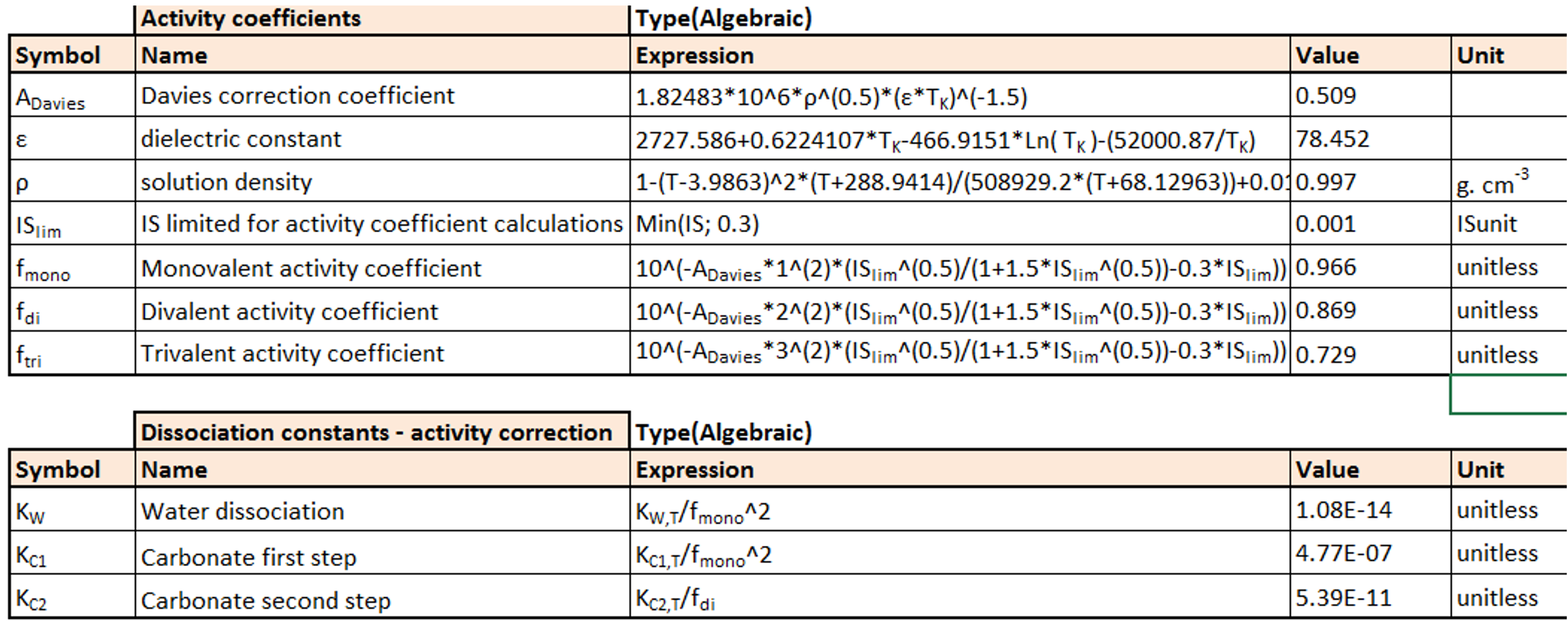
The values for these corrections coefficients are first calculated based on the ionic strength guess.
¶ Speciation calculation
Now the dissociation and activity constants are corrected based on ionic strength guess, the speciation of the components can be calculated:
[H+] concentration is 1e-7 mol/L, kW at 25°C is 1.08e-14, therefore
[OH-] = 1e-14/1e-7 = 1.08e-7
pK of ammonia is 9.244, that is KiNH3 is 1e-9.244.
Let us take total ammonia (SNHx) concentration as 1.4 mgN/L = 0.1 mmol/L = 0.0001 mol/L
[NH3] = 0.0001 * 1e-9.244 /(1.08e-7 + 1e-9.244) = 5.64273e-7 mol/L
[NH4+] = 0.0001 * 1.08e-7 /(1.08e-7 + 1e-9.244) = 9.93879e-5 mol/L (that is at pH 7 most ammonia is ionized)
Those calculations can be found on the pH sheet:

¶ Charge balance, ionic strength and alkalinity calculation
Knowing this speciation, the total charge balance error and the ionic strength can be calculated. On the pH sheet, the charge balance and ionic strength table contains the appropriate number of charges for the charge balance calculation, and the appropriate coefficient to calculate the ionic strength, being calculated as follow:
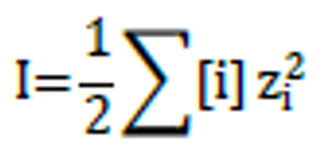
where zi is the charge of the species.
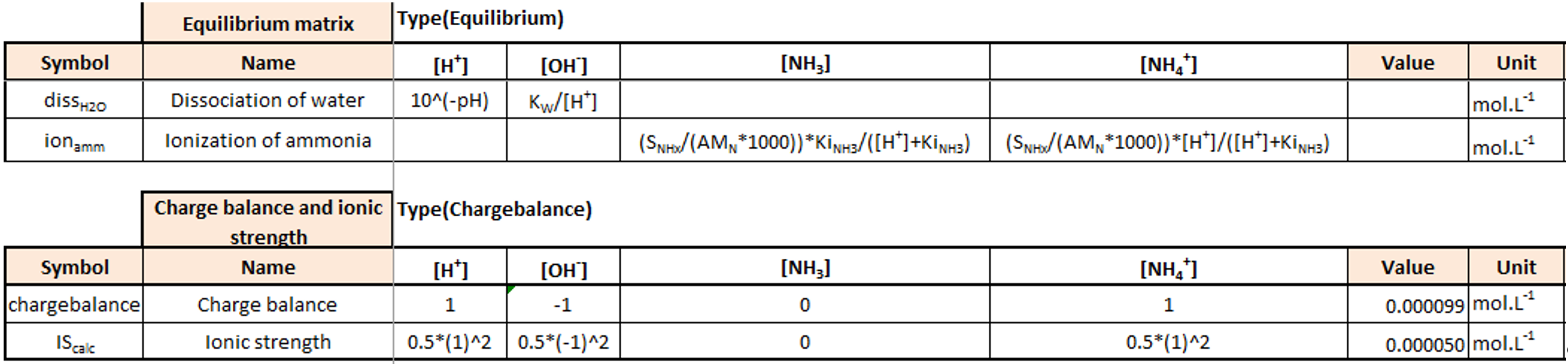
¶ Charge balance calculation
The concentration of each species is multiplied by its corresponding charge, which are summed for all species to calculate the global charge balance. Here, we should have:
Charge balance: [H+] + [NH4+] = [OH-]
¶ Ionic strength calculation
The concentration of each species is multiplied by its corresponding ionic strength coefficient (0.5*zi²), which are summed for all species to calculate the total ionic strength.
¶ Alkalinity calculation
Alkalinity is essentially the amount of monovalent acid we have to add to bring the pH to the equivalence point, about pH 4.1. In this simple example it will be essentially the hydroxyl ion concentration (minus the insignificant proton concentration), adjusted by the free ammonia which will (for practical purposes) all be ionized at pH 4.
SALK = [OH-] + [NH3] - [H+]
These calculations can be found in the Sumo models on the calculated variables tab for the complete system we deal with, plus the chemical matrix on the pH tab.
¶ Resolution of charge balance, ionic strength and alkalinity
As described in the paragraph above, these three variables and the pH are all correlated thus all cannot be set by the user.
On the ammonia solution example, the charge balance calculation is:
[H+] + [NH4+] = [OH-] = 1e-7 + 9.93879e-5
This does not equal to 1.08e-7 so we have a charge balance error (charge balance = 0.000099 mol/L), the pH of a 0.1 mmol/L ammonia solution is not 7. Furthermore, the calculated ionic strength is 0.00005 mol/L, which is different from our guess (0.00001 mol/L).
The task of the Newton-Raphson solver is to solve this system, depending on the options chosen by the user (for influent only, in other process units the second case is used):
| Rules | pH Specification (option available in configure mode for influents) |
Input variables | Variables solved | Targets |
|---|---|---|---|---|
| pHSet | Input pH and alkalinity | pH SALK |
SCAT,NET IS SCO2 |
Chargebalanceerror ISerror SCO2error |
| NonpHSet | Input ions and CO2 | SCO2 SCAT SAN |
pH IS |
Chargebalanceerror ISerror |
¶ Input pH and alkalinity (pH Set)
If the pH 7 is set by the user and a low alkalinity (0.5 mg CaCO3/L) is set, then a charge balance, alkalinity and IS error means that some ions are missing in the solutions. The Newton-Raphson solver will adjust CO2 concentration and calculate the net missing cations until all the errors are close to zero. Then the initial cation or anion concentration is adjusted with the calculated net missing cations as described in the following table:
| Cations and anions calculation revised | Type(Equilibrium) | |
| Symbol | Name | Expression |
| SCAT | Cations concentration | If((SCAT_0 + SCAT,NET) > 0; SCAT_0 + SCAT,NET; SCAT_0) |
| SAN | Anions concentration | If((SCAT_0 + SCAT,NET) > 0; SAN_0; SAN_0 - SCAT,NET) |
On this example the calculated SCO2 concentration is 0.53 g TIC.m-3 and the SCAT,NET calculated is -0.09 mmol.L-1 to counterbalance the ammonium. This leads to the following results:
| Name | Value | Unit |
|---|---|---|
| IS | 9.97E-05 | ISunit |
| SCO2 | 0.53 | mg TIC/L |
| SALK | 0.5 | mg CaCO3/L |
| SCAT_NET | -0.090 | mmol/L |
| SCAT | 0.00 | mmol/L |
| SAN | 0.090 | mmol/L |
| SNHx | 1.4 | g N/m3 |
| [NH4+] | 0.100 | mmol/L |
| [NH3] | 0.00039 | mmol/L |
| chargebalanceerror | 4E-15 | eq.L-1 |
| ISerror | 2E-15 | ISunit |
| SCO2error | -2E-10 | mg CaCO3/L |
¶ Input ions and CO2 (NonpHSet)
All the ionic composition is defined by the user. The Newton-Raphson solver will adjust pH and IS until charge balance error and IS error are close to zero.
The actual pH, with proper molar mass of N, pK values and ionic strength correction in Sumo is 9.79 in this unbuffered solution at 20°C. This leads to the following results:
| Name | Value | Unit |
|---|---|---|
| pH | 9.79 | pHunit |
| IS | 2.89E-05 | ISunit |
| SCO2 | 0 | mg TIC/L |
| SALK | 5.0 | mg CaCO3/L |
| SCAT | 0.000 | mmol/L |
| SAN | 0.000 | mmol/L |
| SNHx | 1.4 | g N/m3 |
| [NH4+] | 0.029 | mmol/L |
| [NH3] | 0.071 | mmol/L |
| chargebalanceerror | -2E-13 | eq.L-1 |
| ISerror | -2E-13 | ISunit |
| SCO2error | nd | mg CaCO3/L |
Musvoto, E., Wentzel, M., Loewenthal, R., Ekama, G., 2000. Integrated chemical-physical processes modelling - I. Development of a kinetic-based model for mixed weak acid/base systems. Water Res. 34, 1857–1867. doi:10.1016/S0043-1354(99)00334-6
Tait, S., Solon, K., Volcke, E.I.P., Batstone, D.J., 2012. A unified approach to modelling wastewater chemistry: model corrections, in: Conference Proceedings: 3rd IWA/WEF Wastewater Treatment Modelling Seminar, WWTmod 2012. Presented at the 3rd Wastewater Treatment Modelling Seminar (WWTmod2012), pp. 51–62.
¶ Chemical phosphorus removal with metal salts addition (iron or aluminium)
The precipitation of amorphous ferric oxide (HFO, considered to be Fe(OH)3(s) by Smith et al. (2011)) provides a number of adsorption sites for ions on its surface, which allow both adsorption and co-precipitation of ions with HFO. The model concepts are developed focusing only on phosphates adsorption and co-precipitation.
The model is based on the concept of reactive site density (named active site factor, ASF expressed as sites/mole of HFO), consisting of oxygen binding sites on HFO for which phosphate species and protons are competing. An equilibrium model, the Surface Complexation Model has already been developed on this basis by Smith et al. (2011, 2008). The total number of available sites per unit of volume (SiteT in mol.m-3) is defined as the product of the mean number of active site factors of HFO (ASFHFO) and the amount of precipitated HFO (XHFO in mol Fe.L-1). Each site has the ability to form bidentate and monodentate surface complexes with HPO42-, H2PO4- and H3PO4 (MUSIC model, (Hiemstra and VanRiemsdijk, 1996)). Bidentate species means that phosphate species (H2PO4- or H3PO4) are bound to two sites owning two metal atoms. To keep the model simple, only one kind of phosphate (without specification of its speciation) is considered to bind onto HFO. The value of the parameter representing the number of active site factors may thus have to be adjusted compared to those used in Surface Complexation Model.
The mean number of active site factors of HFO (ASFHFO) has been found to depend on mixing intensity and HFO aging (Smith et al., 2008; Szabo et al., 2008). Furthermore this equilibrium model was not designed to describe the experimental results observed by Szabo et al. (2008), such as kinetic behaviour of phosphorus removal consisting of an initial fast removal followed by slow removal, and the influence of HFO aging (loss of active surface sites). A detailed dynamic physicochemical model for chemical phosphorus removal with HFO was developed by Hauduc et al (2015). This model is based on the equilibrium model from Smith et al. (2008).
The kinetic model developed in Sumo aims to catch this dynamic behaviour, while keeping the model as simple as possible. The modelling concepts are schematically synthesized on Figure 3 and detailed in the following paragraphs for the main processes considered:
- HFO precipitation
- Phosphorus binding on HFO
- HFO aging
- Phosphorus desorption
- Phosphorus dissolution
¶ HFO precipitation
To simplify the model, the kinetic of HFO precipitation is not considered and aluminium is considered to enter in the reactors of the treatment process directly in the form of HFO. However, the mixing conditions at the dosage point highly impact the size of the flocs. Well mixed metal dosage point will result in smaller flocs. Smaller flocs have higher active site factors, meaning a higher reactive surface compare to the size.
To represent the flocs populations, the model includes 3 flocs types:
- Active hydrous aluminium oxide, high surface (HFO,H) with an active site factor of iASF,HFO,H
- Active hydrous aluminium oxide, low surface (HFO,L) with an active site factor of iASF,HFO,H
- Aged hydrous aluminium oxide (HFO,old) with no active sites
Consequently, the split of the iron dose between the different HFO is considered in the Influent process unit code:
| Symbol | Name | Expression | Unit |
| XHFO,H | Active hydrous ferric oxide, high surface (HFO,H) | G/(KG+G) | g Fe.m-3 |
| XHFO,L | Active hydrous ferric oxide, low surface (HFO,L) | SFe-XHFO,H | g Fe.m-3 |
Table 1 Calculation of HFO,H and HFO,L in Influent process unit code (see definition of parameters in Table 2)
The concentration of active hydrous ferric oxide depends on the impact of mixing intensity, considered through the Monod-type factor: G/(KG+G), with G the average velocity gradient in mixing tank (in s-1) and KG the half saturation coefficient for G value. This factor has a value between 0 and 1 and has a value close to 1 in well mixed conditions.
The parameters for influent process units are the following:
| Symbol | Name | Default | Unit |
| SFe_M | Fe(OH)3 | 0.0 | mg.L-1 |
| G | Average velocity gradient in mixing tank | 50 | s-1 |
| KG | Half saturation coefficient for G value | 10 | s-1 |
Table 2 Influent parameters for aluminium and flocculent dosage
¶ Phosphorus binding on HFO
Phosphate is considered to bound on HFO. The stoichiometric coefficients of the binding processes can be found in Table 3 and Table 3 Stoichiometry of HFO aging processes (with THFO,H = XHFO,H + XHFO,H,P and THFO,L = XHFO,L + XHFO,L,P):
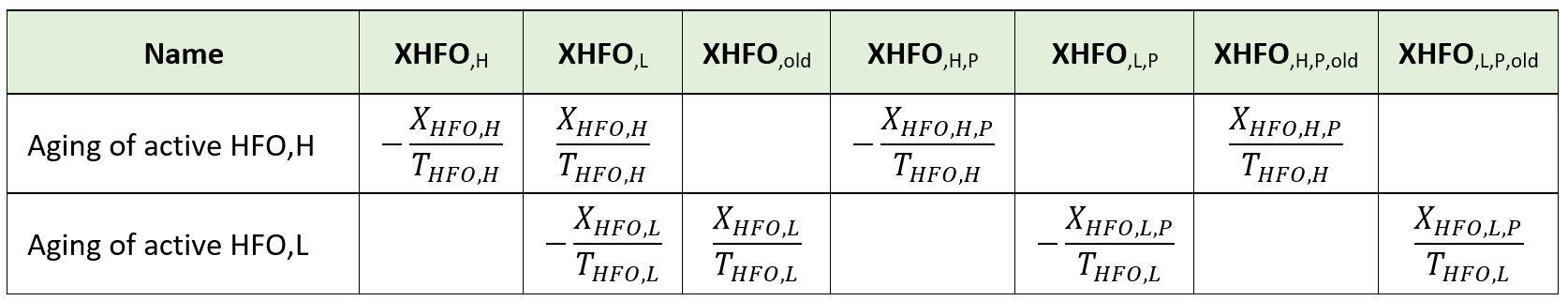
Table 4 with the stoichiometric parameters listed in Table 7, and the kinetic rate expression can be found in Table 6 with the kinetic parameters listed in Table 8.
The model considers the following processes:
- Fast binding of P on active HFO,H to form HFO,H,P
- Slow binding of P on active HFO,L to form HFO,L,P
The kinetic rate expressions are first order to the concentration of free binding sites and a Monod saturation term on the involved phosphorus component ensures the mathematical stability of the model.
¶ Phosphorus desorption
Phosphorus desorption processes are used in the model to reproduce the observed equilibrium between bound and free phosphorus (Smith et al., 2011, 2008). The stoichiometric coefficient can be found in Table 4 with the stoichiometric parameters listed in Table 7, and the kinetic rate expression can be found in Table 6 with the kinetic parameters listed in Table 8. The processes are the reverse of the binding processes:
- Desorption of P on active HFO,H,P to form HFO,H and free phosphate
- Desorption of P on active HFO,L,P to form HFO,L and free phosphate
The kinetic rate expressions are first order to the concentration of each used HFO component and a Monod saturation term on the involved phosphorus component bound on HFO ensures the mathematical stability of the model.
¶ HFO aging
As the model consider only 3 HFO types (High, Low and old). HFO flocs with or without bound phosphate are considered to have the same aging kinetic. Consequently, only 2 aging processes are required, but each includes 3 conversions:
- Fast binding of P on active HFO,H:
| HFO,H => HFO,L | non-used active hydrous aluminium oxide, high surface is turned into active hydrous aluminium oxide, low surface |
| HFO,H,P=>HFO,H,P,old | P-bound hydrous aluminium oxide, high surface is turned into aged P-bound hydrous aluminium oxide, high surface |
The kinetic rate expression is first order to the total HFO,H components (XHFO,H+XHFO,H,P)
- Slow binding of P on active HFO,L
| HFO,L => HFO,old | non-used active hydrous aluminium oxide, low surface is turned into aged unused hydrous aluminium oxide |
|---|---|
| HFO,L,P=>HFO,L,P,old | P-bound hydrous aluminium oxide, low surface is turned into aged P-bound hydrous aluminium oxide, low surface |
The kinetic rate expression is first order to the total HFO,L components (XHFO,L+XHFO,L,P)
Aged used hydrous aluminium oxide are considered inert, meaning that the phosphorus is definitely entrapped in the floc structure, except for HFO,H,P,old and HFO,L,P,old in the phosphorus dissolution process (see next paragraph) for a mathematical purpose.
The stoichiometric coefficient can be found in Table 3 with the stoichiometric parameters listed in Table 7, and the kinetic rate expression can be found in Table 6 with the kinetic parameters listed in Table 8.
¶ Phosphorus dissolution
The phosphorus dissolution process is included in the model to ensure that phosphate will never be depleted for biomass growth. It ensures that below a free phosphate concentration, any bound phosphate on aged HFO will be released, as this bound phosphate cannot be desorbed. The kinetic rate expression is then first order to the total phosphate bound on aged HFO (XHFO,H,P,old+XHFO,L,P,old) and the switch on/off of this process is managed through a logarithm saturation expression that depends on the soluble phosphate concentration.
The stoichiometric coefficient can be found in Table 5 with the stoichiometric parameters listed in Table 7, and the kinetic rate expression can be found in Table 6 with the kinetic parameters listed in Table 8.
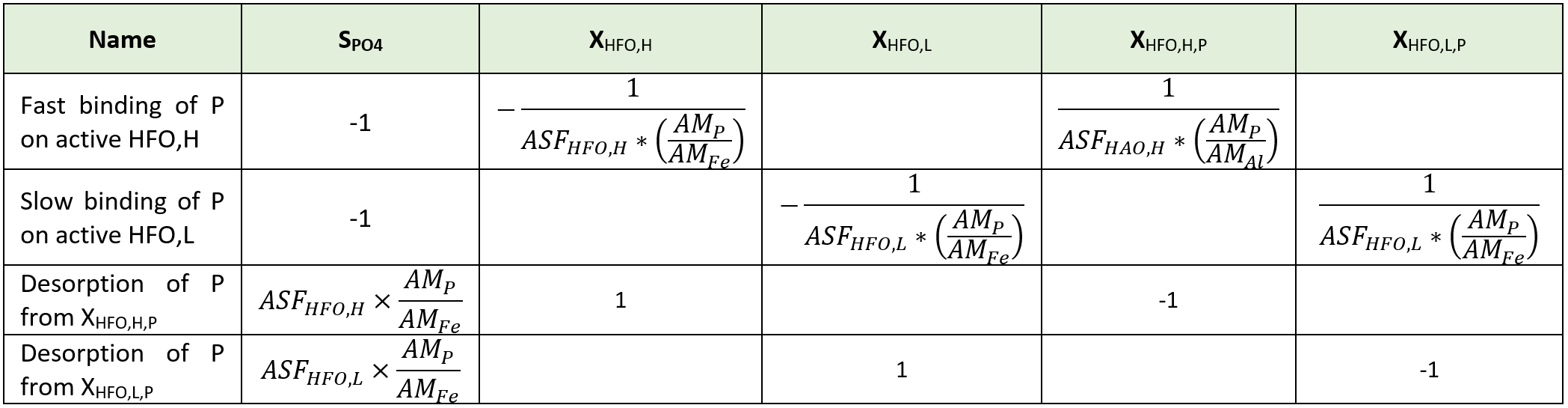
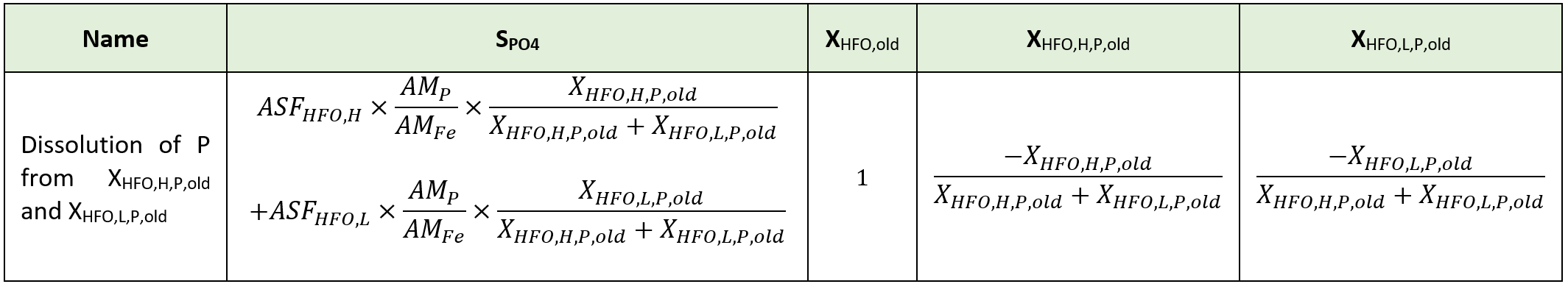
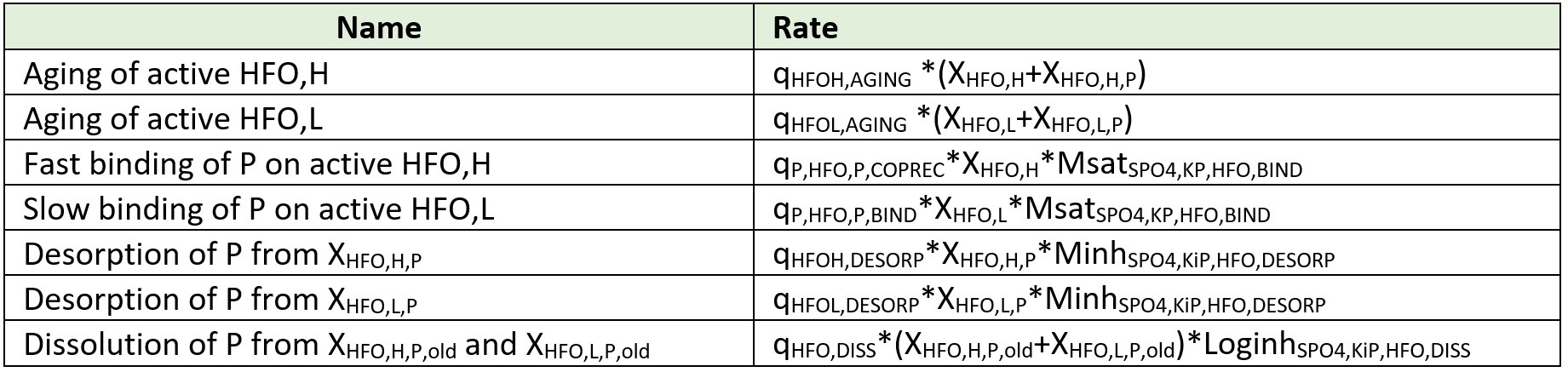
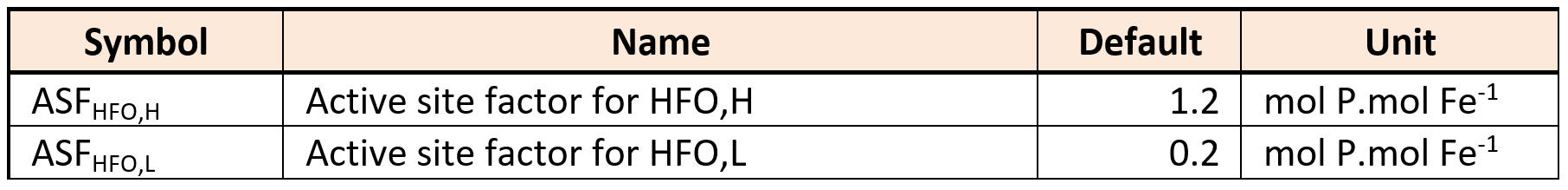
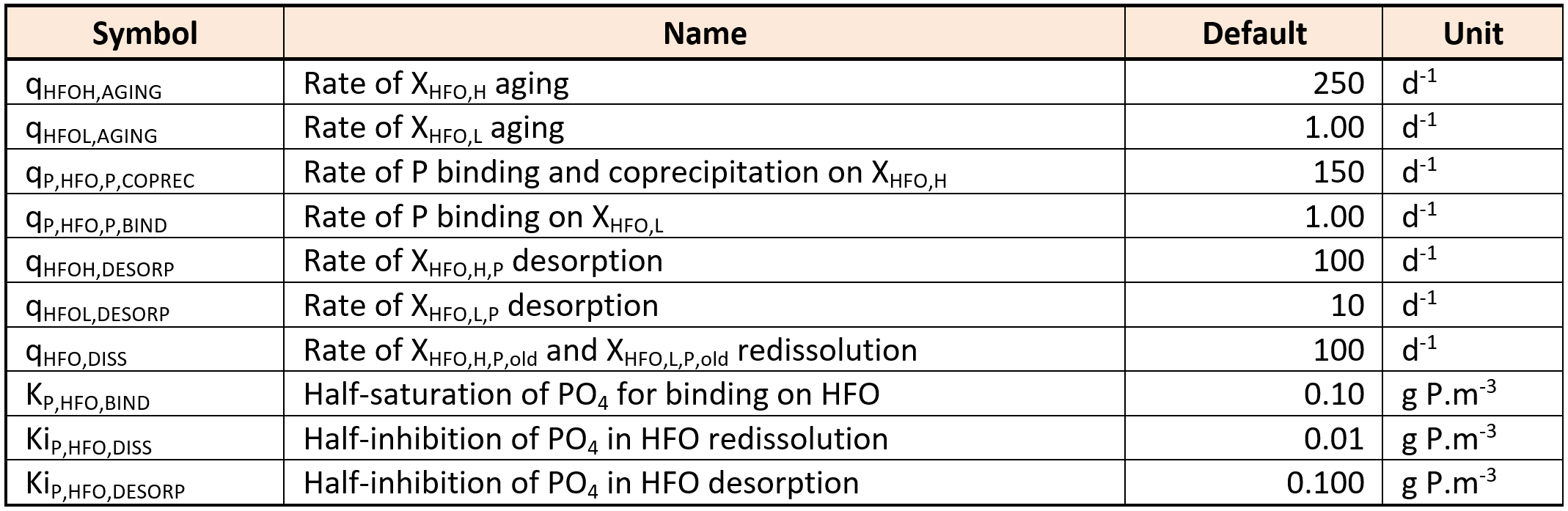
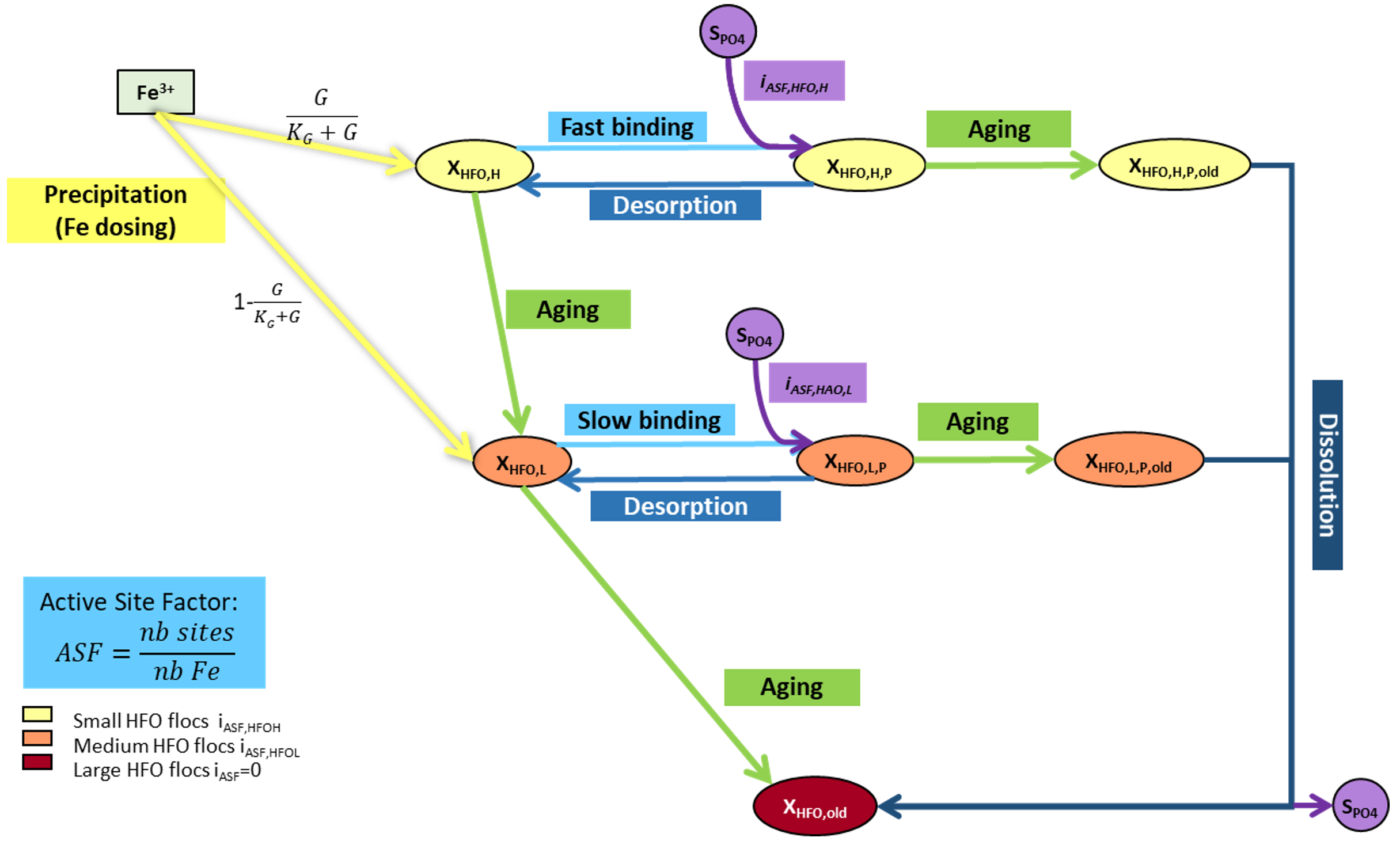
Hauduc, H., Takács, I., Smith, S., Szabo, A., Murthy, S., Daigger, G.T., Spérandio, M., 2015. A dynamic physicochemical model for chemical phosphorus removal. Water Res. 73, 157–170. https://doi.org/10.1016/j.watres.2014.12.053
Hiemstra, T., VanRiemsdijk, W.H., 1996. A surface structural approach to ion adsorption: The charge distribution (CD) model. J. Colloid Interface Sci. 179, 488–508.
https://doi.org/10.1006/jcis.1996.0242
Smith, S., Gray, H., Neethling, J.B., 2011. Surface Complexation Modelling and Aluminum Mediated Phosphorus White Paper | IWA Publishing [WWW Document]. URL https://www.iwapublishing.com/books/9781780406848/surface-complexation-modelling-and-aluminum-mediated-phosphorus-%13-white-paper (accessed 2.26.19).
Smith, S., Takács, I., Murthy, S., Daigger, G.T., Szabo, A., 2008. Phosphate complexation model and its implications for chemical phosphorus removal. Water Environ. Res. 80, 428–438.
Szabo, A., Takács, I., Murthy, S., Daigger, G.T., Licsko, I., Smith, S., 2008. Significance of design and operational variables in chemical phosphorus removal. Water Environ. Res. 80, 407–416. https://doi.org/10.2175/106143008X268498
¶ Chemical precipitation reactions
Six precipitates are implemented in Sumo models (Iron sulfide only available in Sumo2S).
| Symbol | Name | Formula |
|---|---|---|
| XCaCO3 | Calcium carbonate (CaCO3) | CaCO3 |
| XSTR | Struvite (STR) | MgNH4PO4 * 6H2O |
| XBSH | Brushite | CaHPO4 * 2H2O |
| XACP | Amorphous calcium phosphate (ACP) | Ca3(PO4)2 * 4H2O |
| XVivi | Vivianite (Vivi) | Fe3(PO4)2 * 8H2O |
| XFeS | Iron sulfide (FeS) (only in Sumo2S) | FeS |
To predict precipitation, the kinetic rate expression from Musvoto et al. (2000) is used: it depends on a kinetic parameter qPREC and on a driving force that characterizes the deviation from the thermodynamic equilibrium. Mathematically, the driving force is the difference between the ionic product and the solubility constant. The solubility constant is corrected in Sumo for ionic activity, thus the ionic strength effect is modelled.
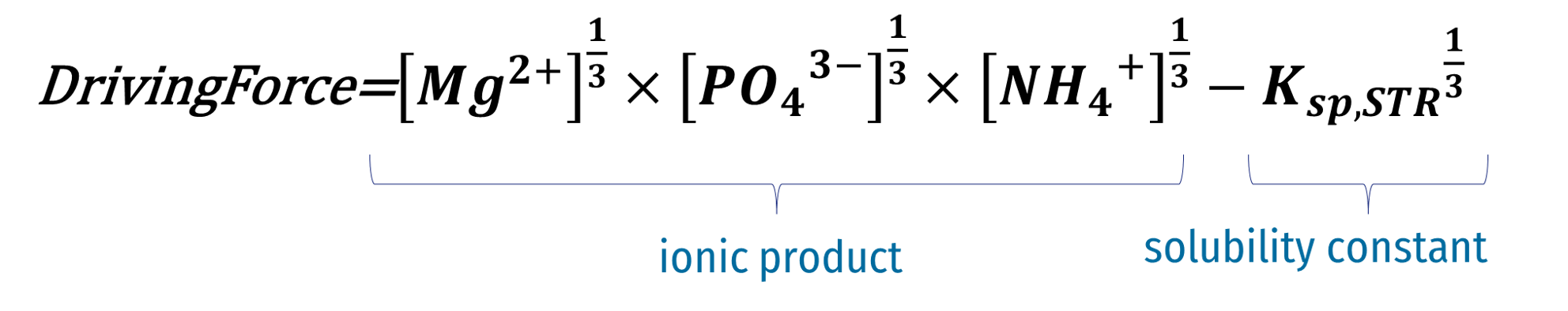
Musvoto, E., Wentzel, M., Loewenthal, R., Ekama, G., 2000. Integrated chemical-physical processes modelling - I. Development of a kinetic-based model for mixed weak acid/base systems. Water Res. 34, 1857e1867. http://dx.doi.org/10.1016/S0043-1354(99)00334-6.
¶ PFAS sorption
Per- and polyfluoroalkyl substances (PFAS) stands for a range of anthropogenic fluorinated compounds used in various applications. These substances do not occur naturally in the environment, they are poorly biodegradable and therefore persistent in nature, accumulating in groundwater, surface water, soil and sediment. A number of PFAS, including the two most commonly present, PFOS (Perfluorooctanesulfonic acid) and PFOA (Perfluorooctanoic acid), have been designated as priority hazardous substances of high concern. Given the negligible biodegradation, the fate and distribution of PFAS in the environment is basically determined by sorption to particulates, which can be described reasonably well as a partitioning-like process.
SUMO24 offers the tracking of PFOA and PFOS by means of a kinetic partition model that you can enable as a model opion in any of the in-house full-plant and focus models. Just simply go to the Models tab and below “Micropollutants”, select the “PFAS are considered” option (by the default setting, they are not considered). This will introduce four additional state variables (particulate and dissolved PFOA and PFOS) and two additional processes (sorption of PFOA and PFOS), along with four additional model parameters (PFOA and PFOS sorption rates and equilibrium distribution coefficients, placed among “Conversion kinetics”).

With special regard to wastewater treatment, median concentrations measured in sewage sludge in relation to median concentrations in effluent suggest that effective water/organic carbon distribution coefficients are to be found around 5000 L/kg for PFOA and 20000 L/kg for PFOS ("global average" values of logKoc were reported by Zareitalabad et al., 2013, as 3.7 and 4.2 for PFOA and PFOS, respectively). These Koc values are used by SUMO as default parameters for the equilibrium distribution coefficients. The kinetics of the sorption mechanism vary largely depending on the matrix and most studies published data regarding different types of soils, therefore their application to wastewater requires caution. SUMO uses a default sorption rate value derived from Wei et al., 2017, however, it is advised to review this parameter based on site measurements, if available, or conduct a sensitivity analysis, if such data is not available. The stoichiometry and the rate expression for PFAS related sorption processes in SUMO are presented below.
Zareitalabad, P., Siemens, J., Hamer, M., Amelung, W., 2013. Perfluorooctanoic acid (PFOA) and perfluorooctanesulfonic acid (PFOS) in surface waters, sediments, soils and wastewater – A review on concentrations and distribution coefficients. Chemosphere 91, 725–732. http://dx.doi.org/10.1016/j.chemosphere.2013.02.024
Wei, C., Song, X., Wang, Q., Hu, Z, 2017. Sorption kinetics, isotherms and mechanisms of PFOS on soils with different physicochemical properties. Ecotoxicology and Environmental Safety 142, 40–50. http://dx.doi.org/10.1016/j.ecoenv.2017.03.040